Probabilistic Model Verification and Validation: A Foundational Framework for Drug Development
This article provides a comprehensive guide to probabilistic model verification and validation, tailored for researchers and professionals in drug development.
Probabilistic Model Verification and Validation: A Foundational Framework for Drug Development
Abstract
This article provides a comprehensive guide to probabilistic model verification and validation, tailored for researchers and professionals in drug development. It explores the foundational principles of probabilistic modeling and its critical role in Model-Informed Drug Development (MIDD). The scope covers a range of methodologies, from quantitative systems pharmacology to AI-driven approaches, and addresses common troubleshooting and optimization challenges. It further details rigorous validation techniques and comparative analyses, synthesizing key takeaways to enhance model reliability, streamline regulatory approval, and accelerate the delivery of new therapies.
Laying the Groundwork: Core Principles of Probabilistic Modeling in Drug Development
Model-Informed Drug Development (MIDD) is an essential framework for advancing drug development and supporting regulatory decision-making by providing quantitative predictions and data-driven insights [1]. Probabilistic models form the backbone of this approach, enabling researchers to quantify uncertainty, variability, and confidence in predictions throughout the drug development lifecycle. These models range from relatively simple quantitative structure-activity relationship (QSAR) models to highly complex quantitative systems pharmacology (QSP) frameworks, all sharing the common goal of informing critical development decisions with mathematical rigor [1] [2]. The fundamental power of MIDD lies in its ability to maximize information from gathered data, build confidence in drug targets and endpoints, and allow for extrapolation to new clinical situations without requiring additional costly studies [2].
The adoption of these probabilistic approaches has transformed modern drug development from a largely empirical process to a more predictive and efficient science. By systematically accounting for variability and uncertainty, these models help accelerate hypothesis testing, assess potential drug candidates more efficiently, reduce costly late-stage failures, and ultimately accelerate market access for patients [1]. Global regulatory agencies now expect drug developers to apply these tools throughout a product's lifecycle where feasible to support key questions for decision-making and validate assumptions to minimize risk [2]. The evolution of these methodologies has been so significant that they have transitioned from "nice to have" components to "regulatory essentials" in late-stage clinical drug development programs [2].
Spectrum of Models in MIDD
The MIDD framework encompasses a diverse set of probabilistic modeling approaches, each with distinct applications, mathematical foundations, and positions along the spectrum from empirical to mechanistic methodologies. Table 1 provides a comparative overview of these key approaches, highlighting their primary applications, probabilistic elements, and regulatory use cases.
Table 1: Key Probabilistic Modeling Approaches in MIDD
| Model Type | Primary Application | Probabilistic Elements | Representative Methods |
|---|---|---|---|
| QSAR [1] | Predict biological activity from chemical structure | Confidence intervals on predictions, uncertainty in descriptor contributions | Regression models, machine learning classifiers |
| PBPK [1] [2] | Predict drug absorption, distribution, metabolism, excretion (ADME) | Inter-individual variability in physiological parameters, uncertainty in system parameters | Virtual population simulations, drug-drug interaction prediction |
| Population PK/PD [1] [2] | Characterize drug exposure and response variability | Random effects for inter- and intra-individual variability, parameter uncertainty | Non-linear mixed effects modeling, covariate analysis |
| QSP [1] [2] | Simulate drug effects on disease pathways | Uncertainty in system parameters, variability in pathway interactions | Virtual patient simulations, disease progression modeling |
| Exposure-Response [1] | Quantify relationship between drug exposure and efficacy/safety | Confidence bands on response curves, prediction intervals | Logistic regression, time-to-event models |
| MBMA [2] | Indirect comparison of treatments across studies | Uncertainty in treatment effect estimates, between-study variability | Hierarchical Bayesian models, meta-regression |
Model Selection Framework
Selecting the appropriate probabilistic model requires careful consideration of the development stage, available data, and specific questions of interest. The "fit-for-purpose" principle dictates that models must be closely aligned with the question of interest (QOI), context of use (COU), and required level of model evaluation [1]. A model is not considered fit-for-purpose when it fails to define the COU, lacks appropriate data quality and quantity, or incorporates unjustified complexities or oversimplifications [1].
The following workflow diagram illustrates the decision process for selecting and applying probabilistic models within the MIDD framework:
Model Selection Workflow in MIDD
This structured approach ensures that model complexity is appropriately matched to the development stage and specific questions being addressed, while maintaining a focus on the regulatory context throughout the process.
Detailed Model Applications and Protocols
Quantitative Structure-Activity Relationship (QSAR) Modeling
QSAR models are computational approaches that predict the biological activity of compounds based on their chemical structure [1]. These models establish probabilistic relationships between molecular descriptors (independent variables) and biological responses (dependent variables), allowing for predictive assessment of novel compounds without synthesis and testing.
Experimental Protocol 1: Development and Validation of a QSAR Model
- Objective: To develop a validated QSAR model for predicting compound activity against a specific biological target.
- Materials:
- Chemical Database: Curated set of compounds with known chemical structures and experimental activity values (e.g., IC50, Ki).
- Descriptor Calculation Software: Tools like RDKit, PaDEL, or Dragon for computing molecular descriptors.
- Statistical Software: R, Python with scikit-learn, or specialized QSAR platforms.
- Procedure:
- Data Curation: Collect and curate a homogeneous dataset of compounds with reliable activity data. Apply stringent exclusion criteria for poor-quality measurements.
- Descriptor Calculation: Compute molecular descriptors (e.g., topological, electronic, geometric) for all compounds in the dataset.
- Descriptor Selection: Apply feature selection techniques (e.g., genetic algorithms, stepwise selection) to reduce dimensionality and avoid overfitting.
- Model Training: Split data into training (≈70-80%) and test (≈20-30%) sets. Apply machine learning algorithms (e.g., partial least squares, random forest, support vector machines) to training data.
- Internal Validation: Assess model performance on training set using cross-validation (e.g., 5-fold or 10-fold) and calculate metrics including Q², R², and root mean square error (RMSE).
- External Validation: Evaluate the final model on the held-out test set to estimate predictive performance on new compounds.
- Applicability Domain: Define the chemical space where the model provides reliable predictions using approaches such as leverage or distance-based methods.
- Quality Assurance: Ensure compliance with OECD principles for QSAR validation, including defined endpoint, unambiguous algorithm, appropriate validation, and applicability domain [3].
Physiologically-Based Pharmacokinetic (PBPK) Modeling
PBPK modeling is a mechanistic approach that simulates how a drug moves through and is processed by different organs and tissues in the body based on physiological, biochemical, and drug-specific properties [2]. These models incorporate probabilistic elements through virtual population simulations that account for inter-individual variability in physiological parameters.
Experimental Protocol 2: PBPK Model Development for Special Populations
- Objective: To develop a PBPK model for predicting drug exposure in unstudied special populations (e.g., pediatrics, hepatic impairment).
- Materials:
- PBPK Software Platform: Commercial (e.g., GastroPlus, Simcyp) or open-source platforms.
- System Data: Population demographic, physiological, and genetic data for target populations.
- Drug-Specific Parameters: In vitro and in vivo data on drug absorption, distribution, metabolism, and excretion.
- Procedure:
- Model Development: Develop and validate a base PBPK model using healthy adult data, incorporating drug-specific parameters (e.g., solubility, permeability, metabolic clearance).
- Sensitivity Analysis: Identify critical system parameters that most influence drug exposure using local or global sensitivity analysis.
- Virtual Population: Generate virtual populations representing the target special population by modifying relevant physiological parameters (e.g., organ sizes, blood flows, enzyme expression).
- Simulation: Execute multiple trials (n=100-1000) with different virtual subjects to predict population pharmacokinetics and account for variability.
- Model Verification: Compare simulation results with any available observed data in the special population, if available.
- Dosing Recommendation: Simulate different dosing regimens to identify optimal dosing for the special population.
- Regulatory Considerations: PBPK models are frequently used to support waivers for clinical studies in special populations and to predict drug-drug interactions [2].
Quantitative Systems Pharmacology (QSP) Modeling
QSP represents the most integrative probabilistic modeling approach, combining computational modeling and experimental data to examine the relationships between a drug, the biological system, and the underlying disease process [4] [2]. These models typically incorporate multiple probabilistic elements, including uncertainty in system parameters and variability in pathway interactions.
Experimental Protocol 3: QSP Model for Combination Therapy Optimization
- Objective: To develop a QSP model for identifying optimal drug combinations in oncology.
- Materials:
- Pathway Data: Literature-curated information on disease-relevant biological pathways.
- Drug Properties: Pharmacokinetic and pharmacodynamic parameters for individual drugs.
- QSP Platform: Specialized software for complex systems modeling.
- Procedure:
- Network Construction: Map key biological pathways and interactions relevant to the disease and drug mechanisms.
- Mathematical Representation: Translate biological network into ordinary differential equations or agent-based models.
- Parameter Estimation: Calibrate model parameters using available preclinical and clinical data, quantifying uncertainty through Bayesian inference or profile likelihood approaches.
- Virtual Patient Generation: Create populations of virtual patients with variability in key biological parameters to represent heterogeneous patient populations.
- Intervention Testing: Simulate monotherapies and combination therapies across the virtual population to identify synergistic effects.
- Biomarker Identification: Simulate potential biomarkers for patient stratification and treatment response prediction.
- Application: QSP is particularly valuable for new modalities, dose selection, combination therapy optimization, and target selection [2].
Probabilistic Aspects and Validation Framework
Quantifying Uncertainty and Variability
A fundamental strength of probabilistic models in MIDD is their explicit handling of two distinct types of randomness: uncertainty and variability. Uncertainty represents limited knowledge about model parameters that could theoretically be reduced with more data, while variability reflects true heterogeneity in populations that cannot be reduced with additional sampling [3].
Table 2 outlines common probabilistic elements and validation approaches across MIDD methodologies:
Table 2: Probabilistic Elements and Validation in MIDD Models
| Model Type | Sources of Uncertainty | Sources of Variability | Validation Approaches |
|---|---|---|---|
| QSAR [1] [3] | Descriptor selection, model structure, activity measurement error | Chemical space diversity, assay variability | External validation, y-randomization, applicability domain assessment |
| PBPK [2] | System parameters, drug-specific parameters, system structure | Physiological differences, enzyme expression, demographics | Prospective prediction vs. observed data, drug-drug interaction verification |
| Population PK/PD [1] [2] | Structural model, parameter estimates, residual error model | Between-subject variability, between-occasion variability | Visual predictive checks, bootstrap analysis, normalized prediction distribution errors |
| QSP [4] [2] | Pathway topology, system parameters, connection strengths | Biological pathway expression, disease heterogeneity | Multilevel validation (molecular, cellular, clinical), prospective prediction |
Model Verification and Validation (V&V) Protocol
Robust validation is essential for establishing confidence in probabilistic models and ensuring their appropriate use in regulatory decision-making. The following protocol provides a general framework for model V&V:
Experimental Protocol 4: Comprehensive Model Verification and Validation
- Objective: To establish a comprehensive V&V framework for probabilistic models in MIDD.
- Procedure:
- Verification:
- Confirm correct implementation of mathematical equations and algorithms.
- Perform unit testing of individual model components.
- Verify numerical accuracy and stability across expected operating ranges.
- Internal Validation:
- Assess model fit to the data used for model development.
- Perform cross-validation (e.g., k-fold, leave-one-out) to evaluate overfitting.
- Conduct sensitivity analysis to identify influential parameters and assumptions.
- External Validation:
- Evaluate model performance against data not used in model development.
- Compare predictions with prospective experimental or clinical results.
- Assess predictive performance using appropriate metrics (e.g., mean absolute error, prediction intervals).
- Predictive Check:
- Generate posterior predictive distributions for key endpoints.
- Compare simulated outcomes with observed data using visual predictive checks.
- Quantify calibration using statistical measures (e.g., prediction-corrected visual predictive checks).
- Verification:
- Documentation: Maintain comprehensive documentation of all model assumptions, data sources, software tools, and validation results to support regulatory submissions [3].
Essential Research Reagents and Computational Tools
Successful implementation of probabilistic models in MIDD requires both computational tools and well-characterized data resources. The following table details key components of the MIDD research toolkit:
Table 3: Research Reagent Solutions for Probabilistic Modeling in MIDD
| Tool Category | Specific Tools/Resources | Function | Key Features |
|---|---|---|---|
| Modeling & Simulation Platforms [1] [2] | NONMEM, Monolix, Simcyp, GastroPlus, R, Python | Implement and execute probabilistic models | Population modeling, PBPK simulation, statistical analysis, machine learning |
| Data Curation Resources [3] [2] | Clinical trial databases, literature compendia, in-house assay data | Provide input data for model development and validation | Highly curated clinical data, standardized assay results, quality-controlled datasets |
| Model Validation Tools [3] | R packages (e.g., xpose, Pirana), Python libraries | Perform model verification, validation, and diagnostic testing | Visual predictive checks, bootstrap analysis, sensitivity analysis |
| Visualization & Reporting [3] | R Shiny, Spotfire, Jupyter Notebooks | Communicate model results and insights to stakeholders | Interactive dashboards, reproducible reports, publication-quality graphics |
Regulatory Context and Future Directions
The regulatory landscape for MIDD approaches has evolved significantly, with global regulatory agencies now encouraging the integration of these approaches into drug submissions [2]. The International Council for Harmonisation (ICH) has developed the M15 guideline to establish global best practices for planning, evaluating, and documenting models in regulatory submissions [4]. This standardization promises to improve consistency among global sponsors in applying MIDD in drug development and regulatory interactions [1].
The FDA's Innovative Science and Technology Approaches for New Drugs (ISTAND) pilot program represents another significant regulatory advancement, designed to qualify novel drug development tools—including M&S and AI-based methods—as regulatory methodologies [4]. These developments, coupled with the FDA's commitment to reducing animal testing through alternatives like MIDD, highlight the growing importance of probabilistic modeling in regulatory science [2].
Looking forward, the integration of artificial intelligence and machine learning with traditional MIDD approaches promises to further enhance their predictive power and efficiency [1] [4]. As these technologies mature, probabilistic models will likely play an increasingly central role in accelerating the development of safer, more effective therapies while reducing costs and animal testing throughout the drug development lifecycle.
The Critical Role of Verification and Validation in Regulatory Success
Verification and validation (V&V) are independent procedures used together to ensure that a product, service, or system meets specified requirements and fulfills its intended purpose [5]. These processes serve as critical components of a quality management system and are fundamental to regulatory success in highly regulated industries such as medical devices and pharmaceuticals. While sometimes used interchangeably, these terms have distinct definitions according to standards adopted by leading organizations [5]. The Institute of Electrical and Electronics Engineers (IEEE) defines validation as "the assurance that a product, service, or system meets the needs of the customer and other identified stakeholders," while verification is "the evaluation of whether or not a product, service, or system complies with a regulation, requirement, specification, or imposed condition" [5]. Similarly, the FDA provides specific definitions for medical devices, stating that validation ensures the device meets user needs and requirements, while verification ensures it meets specified design requirements [5].
A commonly expressed distinction is that validation answers "Are you building the right thing?" while verification answers "Are you building it right?" [5]. This distinction is crucial in regulatory contexts, where a product might pass verification (meeting all specifications) but fail validation (not addressing user needs adequately) if specifications themselves are flawed [5]. For computational models used in regulatory submissions, the ASME V&V 40-2018 standard provides a risk-informed credibility assessment framework that integrates both processes [6].
Theoretical Framework: A Probabilistic Approach to V&V
The probabilistic approach to verification and validation represents a paradigm shift from deterministic checklists to risk-informed, quantitative assessments of model credibility. This approach acknowledges the inherent uncertainties in computational models and provides a framework for quantifying confidence in model predictions, which is particularly valuable for regulatory decision-making [6].
Foundational Concepts
In probabilistic V&V, the traditional binary pass/fail outcome is replaced with a credibility assessment that evaluates the degree of confidence in the model's predictions for a specific Context of Use (COU). The ASME V&V 40 standard establishes a risk-informed process that begins with identifying the question of interest, which describes the specific question, decision, or concern being addressed with a computational model [6]. The COU then establishes the specific role and scope of the model in addressing this question, detailing how model outputs will inform the decision alongside other evidence sources [6].
Risk-Informed Credibility Assessment
The probabilistic framework introduces model risk as a combination of model influence (the contribution of the computational model to the decision relative to other evidence) and decision consequence (the impact of an incorrect decision based on the model) [6]. This risk analysis directly informs the rigor required in V&V activities, with higher-risk applications necessitating more extensive evidence of model credibility [6].
Table: Risk Matrix for Credibility Assessment Planning
| Low Decision Consequence | Medium Decision Consequence | High Decision Consequence | |
|---|---|---|---|
| Low Influence | Minimal V&V Rigor | Moderate V&V Rigor | Substantial V&V Rigor |
| Medium Influence | Moderate V&V Rigor | Substantial V&V Rigor | Extensive V&V Rigor |
| High Influence | Substantial V&V Rigor | Extensive V&V Rigor | Extensive V&V Rigor |
Application Notes: V&V in Regulatory Contexts
Regulatory Landscape for In Silico Evidence
Regulatory agencies increasingly accept evidence produced in silico (through modeling and simulation) to support marketing authorization requests for medical products [6]. The FDA Center for Devices and Radiological Health (CDRH) published guidance on "Reporting of Computational Modeling Studies in Medical Device Submissions" in 2016, followed by the ASME V&V 40-2018 standard in 2018 [6]. Similarly, the European Medicines Agency (EMA) has published guidelines on physiologically based pharmacokinetic (PBPK) modeling, sharing key features with the ASME standard [6].
The Comprehensive in vitro Proarrhythmia Assay (CiPA) initiative represents a significant advancement in regulatory science, proposing modeling and simulation of human ventricular electrophysiology for safety assessment of new pharmaceutical compounds [6]. This initiative, sponsored by FDA, the Cardiac Safety Research Consortium, and the Health and Environmental Science Institute, exemplifies the growing regulatory acceptance of in silico methods when supported by rigorous V&V.
V&V Activities and Methodologies
Verification and validation encompass distinct but complementary activities throughout the development lifecycle:
Verification Activities involve checking that a product, service, or system meets a set of design specifications [5]. In development phases, verification procedures involve special tests to model or simulate portions of the system, followed by analysis of results [5]. In post-development phases, verification involves regularly repeating tests to ensure continued compliance with initial requirements [5]. For machinery and equipment, verification typically consists of:
- Design Qualification (DQ): Confirming through review and testing that equipment meets written acquisition specifications
- Installation Qualification (IQ): Verifying proper installation
- Operational Qualification (OQ): Ensuring operational performance meets specifications
- Performance Qualification (PQ): Demonstrating consistent performance under routine operations [5]
Validation Activities ensure that products, services, or systems meet the operational needs of the user [5]. Validation can be categorized by:
- Prospective validation: Conducted before new items are released
- Retrospective validation: For items already in use, based on historical data
- Partial validation: For research and pilot studies with time constraints
- Re-validation: Conducted after changes, relocation, or specified time periods [5]
Table: Analytical Method Validation Attributes
| Attribute | Description | Application in Probabilistic Framework |
|---|---|---|
| Accuracy and Precision | Closeness to true value and repeatability | Quantified through uncertainty distributions |
| Sensitivity and Specificity | Ability to detect true positives and negatives | Incorporated into model reliability estimates |
| Limit of Detection/Quantification | Lowest detectable/quantifiable amount | Modeled as probability distributions |
| Repeatability/Reproducibility | Consistency under same/different conditions | Source of uncertainty in model predictions |
| Linearity and Range | Proportionality and operating range | Defined with confidence intervals |
Experimental Protocols
Protocol 1: Model Verification Process
Objective: To ensure computational models are implemented correctly and operate as intended.
Materials and Methods:
- Software Tools: Event-B formal modeling tool, Rodin platform, static code analysis tools [7]
- Input Data: Model specifications, design documents, algorithm descriptions
- Verification Techniques:
- Requirement Reviews: Evaluate requirement documents for completeness, clarity, and testability [8]
- Design Reviews: Systematically examine software design artifacts for logical correctness and alignment with requirements [8]
- Code Reviews: Peer review source code to identify defects and enforce standards [8]
- Static Code Analysis: Use automated tools to analyze source code without execution [8]
- Unit Testing: Execute test cases for individual units or functions [8]
- Formal Verification: Use mathematical proof methods, such as those implemented in Event-B, to verify algorithm correctness [7]
Procedure:
- Analyze model requirements to ensure they are clear, complete, and testable
- Plan verification activities, including techniques, responsible personnel, and timeline
- Prepare verification artifacts (requirement specifications, design documents, source code)
- Execute planned verification activities
- Document all findings, including defects and inconsistencies
- Resolve identified issues through development team collaboration
- Perform re-verification to confirm effective corrections
- Compile verification summary report for stakeholder approval [8]
Protocol 2: Model Validation Process
Objective: To ensure computational models meet stakeholder needs and function as intended in real-world scenarios.
Materials and Methods:
- Validation Environment: Realistic test environment mirroring production conditions [8]
- Reference Data: Experimental data, clinical data, historical performance data [6]
- Validation Techniques:
- Functional Testing: Validate software functions against specified requirements [8]
- Integration Testing: Test interactions between integrated modules [8]
- System Testing: Perform end-to-end testing on fully integrated systems [8]
- User Acceptance Testing (UAT): End users verify software meets their needs [8]
- Performance Testing: Validate responsiveness, stability, and scalability [8]
- Uncertainty Quantification: Characterize and quantify uncertainties in model predictions [6]
Procedure:
- Analyze and validate business and functional requirements against end user needs
- Define validation test plan scope, objectives, and schedules
- Design test scenarios and cases simulating real-world usage
- Set up realistic test environment mirroring production conditions
- Execute validation test cases (functional, system, UAT, performance testing)
- Log and analyze defects, focusing on impact to context of use
- Evaluate model credibility through comparison of predictions with validation data
- Compile validation evidence report assessing suitability for regulatory submission [8] [6]
Protocol 3: Probabilistic Neural Network V&V for Load Balancing
Objective: To verify and validate an Effective Probabilistic Neural Network (EPNN) for load balancing in cloud environments using formal methods.
Materials and Methods:
- Modeling Framework: Effective Probabilistic Neural Network (EPNN) architecture [7]
- Verification Tool: Event-B formal modeling tool with Rodin platform [7]
- Algorithms: Round Robin Assigning Algorithm (RRAA), Data Discovery Algorithm (DDA) [7]
- Validation Metrics: Resource utilization, response time, system reliability [7]
Procedure:
- Formal Modeling: Develop formal model of EPNN algorithm in Event-B tool [7]
- Proof Obligation Generation: Use Rodin tool to construct proof obligations based on algorithm context [7]
- Automated Proof: Execute automated proof generation to verify algorithm correctness [7]
- Manual Proof Refinement: Manually correct events not properly associated with invariants or context [7]
- Model Validation: Validate EPNN performance against load balancing metrics (resource utilization, response time) [7]
- Uncertainty Quantification: Characterize probabilistic uncertainties in neural network predictions [7]
- Credibility Assessment: Evaluate model credibility for specific cloud computing contexts of use [7]
The Scientist's Toolkit: Research Reagent Solutions
Table: Essential Tools for Model Verification and Validation
| Tool/Reagent | Function | Application Context |
|---|---|---|
| Event-B Formal Modeling Tool | Provides platform for formal specification and verification of systems | Algorithm correctness verification through mathematical proof [7] |
| Rodin Platform | Open-source toolset for Event-B with automated proof support | Generation of proof obligations and automated proof techniques [7] |
| Static Code Analysis Tools | Analyze source code for defects without execution | Early detection of coding errors and standards compliance [8] |
| Uncertainty Quantification Framework | Characterize and quantify uncertainties in model predictions | Probabilistic assessment of model reliability [6] |
| Traceability Matrix | Verify requirement coverage throughout development | Ensure all requirements have corresponding test coverage [8] |
| Validation Test Environment | Mirror production conditions for realistic testing | System validation under actual operating conditions [8] |
Verification and validation play a critical role in regulatory success by providing evidence of product safety and efficacy. The probabilistic approach to V&V represents an advanced methodology that quantifies model credibility through risk-informed assessment frameworks. By implementing rigorous V&V protocols aligned with regulatory standards such as ASME V&V 40, researchers and product developers can generate compelling evidence for regulatory submissions while ensuring their products reliably meet user needs. The integration of formal verification methods, comprehensive validation testing, and uncertainty quantification provides a robust foundation for regulatory approval in increasingly complex technological landscapes.
In modern drug development and clinical research, the "fit-for-purpose" approach provides a flexible yet rigorous framework for validating models and biomarker assays, ensuring they are appropriate for their specific intended use rather than holding them to universal, one-size-fits-all standards. This paradigm recognizes that the level of evidence and stringency required for validation depends on the model's role in decision-making and its context of use (COU) [9]. Within a broader research thesis on probabilistic approaches to model verification and validation, the fit-for-purpose principle becomes particularly powerful. It allows for the incorporation of uncertainty quantification and probabilistic reasoning, enabling researchers to build models that more accurately represent real-world biological variability and the inherent uncertainties in prediction.
The foundation of this approach lies in aligning the model's capabilities with the specific "Question of Interest" (QoI) it is designed to address. A model intended for early-stage hypothesis generation requires a different validation stringency than one used to support regulatory submissions for dose selection or patient stratification [10]. The context of use explicitly defines the role and scope of the model, the decisions it will inform, and the population and conditions in which it will be applied, forming the critical basis for all subsequent validation activities [11] [10].
The V3 Framework: Verification, Analytical Validation, and Clinical Validation
A comprehensive framework for establishing that a model or Biometric Monitoring Technology (BioMeT) is fit-for-purpose is the V3 framework, which consists of three foundational components: Verification, Analytical Validation, and Clinical Validation [11]. This framework adapts well-established engineering and clinical development practices to the specific challenges of digital medicine and computational modeling.
Verification is the process of confirming through objective evidence that the model's design outputs correctly implement the specified design inputs. In essence, it answers the question: "Did we build the model correctly according to specifications?" [11] [12]. This involves checking that the code is implemented correctly, the algorithms perform as intended in silico, and the computational components meet their predefined requirements.
Analytical Validation moves the evaluation from the bench to an in-vivo context, assessing the performance of the model's algorithms in translating input data into the intended physiological or clinical metrics. It occurs at the intersection of engineering and clinical expertise and is typically performed by the entity that created the algorithm [11]. For a probabilistic model, this would include validating the accuracy of its uncertainty estimates.
Clinical Validation demonstrates that the model acceptably identifies, measures, or predicts a relevant clinical, biological, or functional state within a specific context of use and a defined population [11]. It answers the critical question: "Did we build the right model for the intended clinical purpose?" [12]. This requires evidence that the model's outputs correlate meaningfully with clinical endpoints or realities.
The relationship and primary questions addressed by these components are summarized in the workflow below.
Quantitative Performance Standards for Method Validation
The specific performance parameters evaluated during validation are highly dependent on the type of model or assay being developed. The fit-for-purpose approach tailors the validation requirements to the assay's technology category and its position on the spectrum from research tool to clinical endpoint. The American Association of Pharmaceutical Scientists (AAPS) has identified five general classes of biomarker assays, each with recommended performance parameters to investigate during validation [9].
Table 1: Recommended Performance Parameters by Assay Category
| Performance Characteristic | Definitive Quantitative | Relative Quantitative | Quasi-Quantitative | Qualitative |
|---|---|---|---|---|
| Accuracy | + | |||
| Trueness (Bias) | + | + | ||
| Precision | + | + | + | |
| Reproducibility | + | |||
| Sensitivity | + | + | + | + |
| Specificity | + | + | + | + |
| Dilution Linearity | + | + | ||
| Parallelism | + | + | ||
| Assay Range | + | + | + | |
| LLOQ/ULOQ | + (LLOQ-ULOQ) | + (LLOQ-ULOQ) |
For definitive quantitative methods (e.g., mass spectrometric analysis), accuracy is dependent on total error, which is the sum of systematic error (bias) and random error (intermediate precision) [9]. While bioanalysis of small molecules traditionally follows the "4-6-15 rule" (where 4 of 6 quality control samples must be within 15% of their nominal value), biomarker method validation often allows for more flexibility, with 25% being a common default value for precision and accuracy (30% at the Lower Limit of Quantitation) [9]. A more sophisticated approach involves constructing an "accuracy profile" which plots the β-expectation tolerance interval to visually display the confidence interval (e.g., 95%) for future measurements, allowing researchers to determine the probability that future results will fall within pre-defined acceptance limits [9].
Experimental Protocols for Fit-for-Purpose Validation
Protocol: Multi-Stage Validation for Biomarker Assays
This protocol outlines a phased approach for biomarker method validation, emphasizing iterative improvement and continuous assessment of fitness-for-purpose [9].
1. Purpose and Goal To establish a robust, phased methodology for the validation of biomarker assays, ensuring they are fit-for-purpose for their specific intended use in clinical trials or research.
2. Experimental Workflow The validation process proceeds through five discrete stages:
Stage 1: Definition and Selection
- Define the explicit purpose and Context of Use (COU) for the assay.
- Select the candidate assay based on the COU and the biological question.
- Output: A clearly articulated validation goal.
Stage 2: Planning and Assembly
- Assemble all necessary reagents, components, and data pipelines.
- Write a detailed method validation plan.
- Finalize the classification of the assay (e.g., definitive quantitative, qualitative).
- Output: A comprehensive validation protocol.
Stage 3: Experimental Performance Verification
- Execute the validation plan to characterize the assay's performance parameters (see Table 1).
- Critically evaluate fitness-for-purpose by comparing performance data against the pre-defined acceptance criteria from Stage 1.
- Output: A validation report and a Standard Operating Procedure (SOP) for routine use.
Stage 4: In-Study Validation
- Deploy the assay in a pilot or actual clinical study.
- Assess robustness in the clinical context and identify real-world issues (e.g., related to patient sample collection, storage, and stability).
- Output: An assessment of practical fitness-for-purpose.
Stage 5: Routine Use and Monitoring
- Implement the assay for its intended routine use.
- Establish ongoing Quality Control (QC) monitoring, proficiency testing, and procedures for handling batch-to-batch QC issues.
- Output: A system for continuous quality assurance and iterative improvement.
3. Key Considerations
- The driver of the process is continuous improvement, which may necessitate iterations that loop back to any earlier stage.
- For probabilistic models, each stage should include steps for evaluating uncertainty calibration, such as calculating metrics like the Expected Calibration Error (ECE) [13].
Protocol: Implementing a Probabilistic Phenotyping Model
This protocol details the methodology for building a probabilistic disease phenotype from Electronic Health Records (EHR) using the Label Estimation via Inference (LEVI) model, a Bayesian approach that does not require gold-standard labels [13].
1. Purpose and Goal To create a probabilistically calibrated disease phenotype from EHR data that outputs well-calibrated probabilities of diagnosis instead of binary classifications, enabling better risk-benefit tradeoffs in downstream applications.
2. Experimental Workflow
Step 1: Candidate Population Filtering
- Filter the EHR population down to disease-specific candidates using broad, inclusive criteria (e.g., presence of relevant diagnosis codes, medications, or terms in clinical notes).
- Goal: Increase disease prevalence in the candidate pool while minimizing the exclusion of true positive cases.
Step 2: Develop Labeling Functions (LFs)
- Iteratively develop a set of heuristic "labeling functions" – rules that vote on a patient's positive or negative status.
- LFs can apply to any data modality (e.g., "patient prescribed drug X," "disease Y mentioned in a numbered list").
- Validate LFs through spot-checking accuracy and clinician consultation.
Step 3: Aggregate Votes Using LEVI Model
- Apply the LEVI model, which computes the posterior probability of a positive diagnosis using a closed-form Bayesian solution. The model leverages:
α_ρ,β_ρ: Priors for disease prevalence.α_TPR,β_TPR: Priors for the True Positive Rate of positive LFs.α_FPR,β_FPR: Priors for the False Positive Rate of positive LFs.
- The posterior label probability is derived as:
P(z_j=1 | V_j) = σ( log( (α_ρ + n_pos - 1)/(β_ρ + n_neg - 1) ) + Σ_i:V_ij=1 log( (α_TPR + k_TP_i - 1)/(α_FPR + k_FP_i - 1) ) + Σ_i:V_ij=0 log( (β_FPR + N_i - k_FP_i - 1)/(β_TPR + N_i - k_TP_i - 1) ) )Whereσis the logistic function,V_jis the vote vector for patientj,n_pos/n_negare counts of positive/negative votes, andk_TP_i/k_FP_iare counts of true/false positives for LFiestimated from the data.
- Apply the LEVI model, which computes the posterior probability of a positive diagnosis using a closed-form Bayesian solution. The model leverages:
Step 4: Prior Selection via Maximum Entropy
- Encode prior knowledge about prevalence and LF performance using the principle of Maximum Entropy, making the prior distribution as non-committal as possible given known constraints (e.g., an "upper bound" on a variable's plausible value).
3. Key Considerations
- This method is particularly valuable in EHR data, which often suffers from incompleteness, leading to ambiguous or contradictory evidence for a diagnosis.
- The output is a well-calibrated probability, which allows for more nuanced decision-making based on the specific costs of false positives and false negatives in a given application [13].
The following diagram illustrates the key stages of this probabilistic phenotyping process.
The Scientist's Toolkit: Essential Research Reagents and Materials
Successfully implementing a fit-for-purpose validation strategy requires a suite of methodological tools and conceptual frameworks. The table below details key "research reagents" essential for this process.
Table 2: Essential Reagents for Fit-for-Purpose Model Validation
| Tool Category | Specific Tool/Technique | Function & Purpose |
|---|---|---|
| Conceptual Framework | Context of Use (COU) | Defines the specific role, scope, and decision-making context of the model, forming the foundation for all validation activities [10]. |
| Conceptual Framework | Question of Interest (QoI) | Articulates the precise scientific or clinical question the model is designed to address, ensuring alignment between the model and its application [10]. |
| Validation Framework | V3 (Verification, Analytical Validation, Clinical Validation) | Provides a structured, three-component framework for the foundational evaluation of models and BioMeTs [11]. |
| Statistical Tool | Accuracy Profile / β-Expectation Tolerance Interval | A visual tool for assessing the total error of a quantitative method, predicting the confidence interval for future measurements against pre-defined acceptance limits [9]. |
| Computational Model | Label Estimation via Inference (LEVI) | A Bayesian model for aggregating weak supervision signals to create probabilistically calibrated outputs without the need for gold-standard labels [13]. |
| Regulatory Document | Model Analysis Plan (MAP) | A pre-defined plan outlining the technical criteria, assumptions, and analysis pipeline for a model, serving as the foundation for regulatory alignment [10]. |
| Quality Control Tool | Traceability Matrix | A document that links requirements (e.g., user needs, design inputs) to corresponding verification and validation activities, ensuring comprehensive coverage [14]. |
Adopting a fit-for-purpose approach is fundamental to developing models that are not only scientifically sound but also clinically meaningful and resource-efficient. By rigorously aligning the model with a specific Question of Interest and Context of Use, and by employing structured frameworks like V3 and probabilistic methodologies like LEVI, researchers can generate the robust evidence base needed to support critical decisions in drug development and clinical practice. This paradigm, especially when integrated with probabilistic reasoning, ensures that models are deployed with a clear understanding of their capabilities, limitations, and inherent uncertainties, ultimately enhancing the reliability and impact of model-informed drug development.
In the rigorous framework of probabilistic model verification and validation, the drug development process represents a critical domain for applying structured uncertainty quantification. Transition probabilities—the quantitative metrics that define the likelihood a drug candidate moves from one clinical phase to the next—serve as fundamental parameters in state-transition models that predict research outcomes, resource allocation, and ultimate commercial viability [15]. These probabilities form the mathematical backbone of cost-effectiveness analyses and portfolio decision-making, translating complex, multi-stage clinical development pathways into computable risk metrics.
Understanding and accurately estimating these probabilities is essential for creating robust models that reflect the actual dynamics of drug development. This overview examines the methodologies for deriving these critical values from published evidence, explores disease-specific variations that challenge aggregate estimates, and provides structured protocols for their application in probabilistic research models, thereby contributing to more reliable verification and validation of developmental risk assessments.
Defining Transition Probabilities in Clinical Development
Transition probabilities are mathematically defined as the probability that a drug product moves from one defined clinical phase to the next during a specified time period, known as the cycle length [15]. In the context of a state-transition model for drug development, these probabilities quantify the risk of progression through sequential stages: typically from Phase I to Phase II, Phase II to Phase III, and Phase III to regulatory approval and launch.
These probabilities are cumulative, representing the compound likelihood of successfully overcoming all scientific, clinical, and regulatory hurdles within a phase. Decision modelers face two primary challenges: published data often comes in forms other than probabilities (e.g., rates, relative risks), and the time frames of published probabilities rarely match the cycle length required for a specific model [15].
The International Society for Pharmacoeconomics and Outcomes Research (ISPOR)–Society for Medical Decision Making (SMDM) Modeling Task Force recommends deriving transition probabilities from "the most representative data sources for the decision problem" [15]. The hierarchy of evidence sources includes:
- Population-based epidemiological studies are preferred for modeling the natural history of a condition.
- Randomized Controlled Trials (RCTs) provide the highest-quality evidence of efficacy for intervention arms, though generalizability to real-world settings can be limited.
- Network meta-analyses offer a robust methodology for comparing multiple interventions that have not been directly tested against each other in single RCTs, maintaining randomization within trials for unbiased relative treatment effects [15].
Table 1: Common Statistical Measures Used to Derive Transition Probabilities
| Statistic | Definition | Range | Use in Probability Derivation |
|---|---|---|---|
| Probability/Risk | Number of events / Number of people followed | 0–1 | Direct input; may require cycle-length adjustment |
| Rate | Number of events / Total person-time experienced | 0 to ∞ | Converted to probability using survival formulas |
| Relative Risk (RR) | Probability in exposed / Probability in unexposed | 0 to ∞ | Adjusts baseline probabilities for subgroups or treatments |
| Odds | Probability / (1 - Probability) | 0 to ∞ | Intermediate step in calculations |
| Odds Ratio (OR) | Odds in exposed / Odds in unexposed | 0 to ∞ | Used to adjust probabilities via logistic models |
Disease-Specific Variations in Transition Probabilities
Aggregate transition probabilities at the therapeutic area level can mask significant variations at the individual disease level, a critical consideration for accurate model validation. Research analyzing eight specific diseases revealed that for five of them, success probabilities for individual diseases deviated meaningfully (by more than ten percentage points) from the broader neurological or autoimmune therapeutic area probabilities [16].
Table 2: Comparative Cumulative Phase Success Probabilities by Disease [16]
| Disease / Therapeutic Area | Phase I to II | Phase II to III | Phase III to Launch |
|---|---|---|---|
| Neurology (Therapeutic Area) | 62% | 19% | 9-15% |
| Amyotrophic Lateral Sclerosis (ALS) | 75% | 27% | 4% |
| Autoimmune (Therapeutic Area) | Data not specified | Data not specified | Data not specified |
| Crohn's Disease | Aligned closely with autoimmune area | Aligned closely with autoimmune area | Aligned closely with autoimmune area |
| Rheumatoid Arthritis (RA) | Aligned closely with autoimmune area | Aligned closely with autoimmune area | Aligned closely with autoimmune area |
| Multiple Sclerosis (MS) | Aligned closely with autoimmune area | Aligned closely with autoimmune area | Aligned closely with autoimmune area |
Key observations from this comparative analysis include:
- Neurological Disease Deviation: For Amyotrophic Lateral Sclerosis (ALS), drugs showed a higher probability of success in early phases (Phase I to II and Phase II to III) compared to the neurological area overall. However, this advantage reversed dramatically in the final phase, where the probability of success from Phase III to launch (4%) was less than half that of the broader neurology area (9-15%) [16].
- Autoimmune Disease Consistency: In contrast, the three autoimmune disorders studied—Crohn's disease, Rheumatoid Arthritis, and Multiple Sclerosis—followed nearly identical success probability trajectories that closely matched their overarching therapeutic area. This consistency occurred despite differences in the availability of efficacy biomarkers, suggesting that other factors beyond biomarker understanding influence phase transition success in this domain [16].
- Rarity and Biomarker Impact: The study found no clear trends linking success probabilities to whether a disease was rare or not. The presence of efficacy biomarkers (classified as high/medium for MS and RA, but not for Crohn's) was noted as important for Phase III success but not determinative, indicating that multiple factors govern transition outcomes [16].
These findings underscore a critical principle for model verification: the use of therapeutic-area-level transition probabilities as precise predictors for specific diseases within that area can be misleading. Effective probabilistic validation must account for this heterogeneity by incorporating disease-specific data where material differences exist.
Methodological Framework and Experimental Protocols
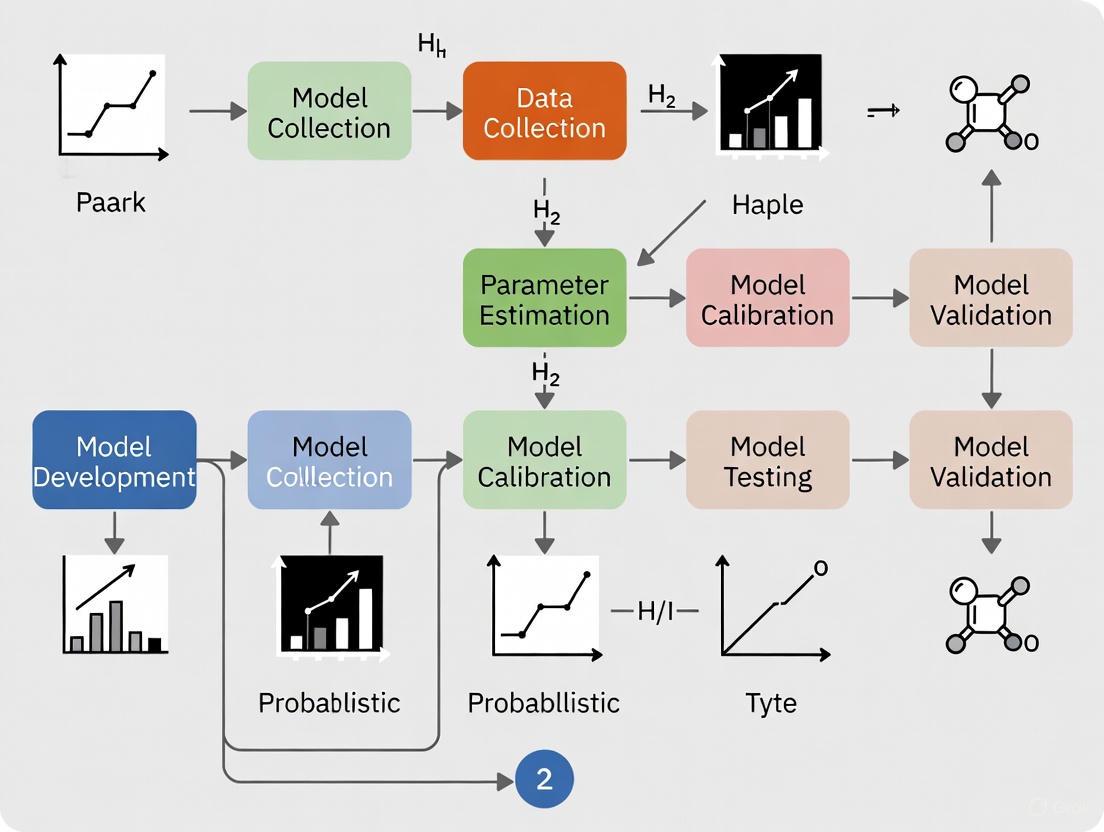
Workflow for Deriving and Applying Transition Probabilities
The following diagram outlines the comprehensive methodology for building a probabilistic drug development model, from data acquisition to validation.
Protocol 1: Deriving Transition Probabilities from Published Statistics
This protocol details the conversion of common published statistics into usable transition probabilities, corresponding to the "Conversion" node in the workflow.
Objective: To transform relative risks, odds ratios, rates, and probabilities with mismatched time frames into cycle-length-specific transition probabilities for state-transition models.
Materials and Inputs:
- Published study reports containing relevant statistics (see Table 1)
- Baseline probability for the target population (for RR and OR conversions)
- Defined cycle length for the decision model
Methodology:
From Relative Risk (RR) to Probability:
- Obtain the relative risk ((RR)) and the baseline probability ((p_{control})) from the literature.
- Calculate the probability in the treated/exposed group as: (p{treatment} = RR \times p{control}).
- Ensure the resulting probability is within the valid range [0,1]; if not, consider using a logistic transformation.
From Odds and Odds Ratios (OR) to Probability:
- If the odds ((O)) is provided, convert to probability using: (p = O / (1 + O)).
- If an odds ratio ((OR)) and baseline probability ((p{control})) are provided:
- First, convert the baseline probability to odds: (O{control} = p{control} / (1 - p{control})).
- Calculate the odds for the treated group: (O{treatment} = OR \times O{control}).
- Convert the resulting odds back to a probability: (p{treatment} = O{treatment} / (1 + O_{treatment})).
From Rates to Probabilities:
- Acquire the event rate ((r)) per unit time.
- Use the exponential transformation to calculate the probability ((p)) over a specific cycle length ((t)): (p = 1 - e^{-r \times t}).
- This formula assumes a constant hazard rate over the interval.
Cycle Length Adjustment (Two-State Model):
- For a known probability ((p)) over a given time period ((T)), and a target cycle length ((t)), the corresponding transition probability ((pt)) can be derived from the rate: (r = -ln(1-p) / T), followed by (pt = 1 - e^{-r \times t}).
- This method is appropriate only when two state transitions are possible (e.g., remaining in the state or moving to one other state).
Validation Steps:
- Cross-validate derived probabilities against any reported probabilities in the source literature.
- Perform unit checks to ensure all probabilities fall between 0 and 1.
- Confirm that probabilities for all transitions from a single state sum to 1 (or ≤1 if censoring is present).
Protocol 2: Incorporating Disease-Specific and Model Structure Adjustments
This protocol addresses the advanced modeling techniques referenced in the "Disease Adjustments" and "Model Populate" nodes of the workflow.
Objective: To adjust aggregate therapeutic-area probabilities for specific diseases and to handle models with three or more potential transitions from a single state.
Materials and Inputs:
- Disease-level clinical trial data (e.g., from Citeline's Pharmaprojects or similar databases) [16]
- Aggregate therapeutic-area transition probabilities
- State-transition model with defined health states
Methodology:
Implementing Disease-Specific Adjustments:
- Identify Material Deviations: Compare available disease-specific success probabilities (see Table 2) to therapeutic-area benchmarks. A deviation of >10 percentage points is suggested as a threshold for "meaningful" difference [16].
- Parameter Replacement: Where material deviations exist, replace the aggregate probability with the disease-specific estimate.
- Stratified Analysis: If data permits, stratify probabilities further by relevant factors such as drug class (e.g., TNF blockers) or the presence of efficacy biomarkers, though their predictive power may vary [16].
Handling Multiple Health-State Transitions:
- Challenge Identification: The standard two-state cycle-length adjustment fails when a patient can transition to three or more states in a single cycle (e.g., from "Local Cancer" to "Regional Cancer," "Metastatic Cancer," or "Death").
- Rate Matrix Approach:
- Define a transition rate for each possible state change.
- Organize these rates into a matrix ((R)).
- Use matrix exponentiation to calculate the transition probabilities over the model's cycle length: (P(t) = e^{R \times t}).
- Bootstrapping as an Alternative: When individual-level patient data is available, bootstrapping can be used to estimate the distribution of transition probabilities for complex multi-state models.
Sensitivity Analysis and Uncertainty Allocation:
- Parameter Uncertainty: Use probabilistic sensitivity analysis (PSA) by assigning probability distributions (e.g., Beta for probabilities, Gamma for rates) to key transition parameters and running Monte Carlo simulations.
- Model Uncertainty: Evaluate the impact of using different data sources (e.g., disease-specific vs. therapeutic-area probabilities) on the model's outcomes.
- Structural Uncertainty: Test alternative model structures, such as different cycle lengths or state definitions, to ensure the robustness of conclusions derived from the transition probabilities.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Resources for Transition Probability Analysis
| Tool / Resource | Type | Primary Function | Application Context |
|---|---|---|---|
| Citeline's Pharmaprojects | Commercial Database | Tracks drug development programmes from start to success/failure; provides disease-level trial data. | Source for deriving disease-specific transition probabilities and analyzing development trends [16]. |
| Network Meta-Analysis | Statistical Methodology | Enables indirect comparison of multiple interventions using Bayesian framework to generate probabilistic outputs. | Generating relative treatment effects and transition probabilities for drugs not directly compared in head-to-head trials [15]. |
| Probabilistic Model Checker | Software Tool | Formally verifies temporal properties of probabilistic models against specified requirements. | Checking safety and liveness properties of state-transition models under uncertainty [17]. |
| State-Transition Model | Modeling Framework | A Markov or semi-Markov model that simulates the progression of cohorts through health states using transition probabilities. | Core structure for cost-effectiveness analysis and drug development risk projection [15]. |
| Monte Carlo Simulation | Computational Algorithm | Randomly samples input distributions (e.g., of transition probabilities) to quantify outcome uncertainty. | Conducting probabilistic sensitivity analysis to understand the impact of parameter uncertainty on model results [15]. |
Transition probabilities are more than mere inputs for clinical development models; they are the fundamental parameters that encode the complex, stochastic reality of drug development into a verifiable and validatable quantitative framework. Their accurate derivation from published evidence—whether from rates, relative risks, or odds ratios—and their proper adjustment for disease-specific contexts are critical steps in building models that truly reflect underlying risks. The methodological protocols and toolkit presented here provide a structured approach for researchers to quantify development risk rigorously. By applying these principles within the broader context of probabilistic verification and validation, modelers can enhance the reliability of their predictions, ultimately supporting more informed and resilient decision-making in pharmaceutical research and development.
Historical Context and the Evolution of Probabilistic Analysis in Biomedical Research
The integration of probabilistic reasoning into biomedical research represents a fundamental paradigm shift from authority-based medicine to evidence-based science. This transition, which began centuries ago, has transformed how researchers quantify therapeutic effectiveness, validate models, and manage uncertainty in clinical decision-making. The historical development of this approach reveals a persistent tension between clinical tradition and mathematical formalization, with key breakthroughs often emerging from interdisciplinary collaboration.
The 18th century marked a crucial turning point, as physicians began moving away from absolute confidence in medical authority toward reliance on relative results based on systematic observation [18]. British naval physician James Lind captured this emerging probabilistic mindset in 1772 when he noted that while "a work more perfect and remedies more absolutely certain might perhaps have been expected from an inspection of several thousand patients," such "certainty was deceitful" because "more enlarged experience must ever evince the fallacy of positive assertions in the healing art" [18]. This recognition of inherent uncertainty in therapeutic outcomes laid the groundwork for more formal statistical approaches.
Historical Foundations: Key Figures and Conceptual Breakthroughs
Early Pioneers of Medical Probability
Table 1: Key Historical Figures in Medical Probabilistic Reasoning
| Figure | Time Period | Contribution | Conceptual Approach |
|---|---|---|---|
| James Lind | 1716-1794 | Systematic observation and reporting of all cases (successes and failures) | Unconscious probabilistic reasoning through complete case reporting [18] |
| John Gregory | 1724-1773 | Explicit use of term "probability" in medical context | Conscious, pre-mathematical probabilistic reasoning [18] |
| John Haygarth | 1740-1824 | Application of mathematical probability to smallpox infection | Conscious, mathematical mode using "doctrine of chances" [18] |
| Carl Liebermeister | 1833-1901 | Probability theory applied to therapeutic statistics | Radical solution to problem of arbitrary statistical thresholds [19] |
The 19th century witnessed further formalization of these approaches. German physician Carl Liebermeister made remarkable contributions with his 1877 paper "Über Wahrscheinlichkeitsrechnung in Anwendung auf therapeutische Statistik" (On Probability Theory Applied to Therapeutic Statistics), which offered innovative solutions to the problem of arbitrary probability thresholds in assessing therapeutic effectiveness [19]. Liebermeister recognized that available statistical theory was "so far been too incomplete and inconvenient" for practical clinical use, and he challenged the prevailing "unshakeable dogma" that "series of observations which do not consist of very large numbers cannot prove anything at all" [19]. His work provided building blocks for a paradigm shift in medical statistics that would have better served clinicians than today's predominant methodology.
The Formalization of Statistical Methods
The early 20th century saw the emergence of frequentist statistics as a dominant paradigm, largely driven by practical considerations. As noted in historical analyses, "prior to the computer age, you had to be a serious mathematician to do a proper Bayesian calculation," but frequentist methods could be implemented using "probability tables in big books that mere mortals such as you or I could pull off the shelf" [20]. This accessibility led to widespread adoption, though not always with proper understanding. By 1929, a review of 200 medical research papers found that 90% should have used statistical methods but didn't, and just three years later, concerns were already being raised about frequent violations of "the fundamental principles of statistical or of general logical reasoning" [20].
Diagram 1: Historical Evolution of Probabilistic Analysis in Biomedical Research
Modern Probabilistic Analysis: Applications and Protocols
Current Applications in Biomedical Research
The life science analytics market, valued at USD 11.27 billion in 2025, reflects the massive adoption of probabilistic and data analytics techniques across biomedical research [21]. This growth is driven by several key applications:
Drug Discovery and Development: Advanced analytics help identify promising drug candidates, predict trial outcomes, and optimize study protocols to reduce time and costs [21]. The integration of diverse data sources, including genomics and real-world evidence, enables more informed decision-making throughout the R&D pipeline.
Clinical Data Science Evolution: Traditional clinical data management is evolving into clinical data science, with professionals shifting from operational tasks (data collection and cleaning) to strategic contributions (generating insights and predicting outcomes) [22]. This transition requires new skill sets and represents a fundamental change in how clinical data is utilized.
Risk-Based Approaches: Regulatory support for risk-based quality management (RBQM) and data management is encouraging sponsors to focus on critical-to-quality factors rather than comprehensive data review [22]. This approach introduces higher data quality through proactive issue detection, greater resource efficiency via centralized data reviews, and shorter study timelines.
Protocol: Model-Based Experimental Manipulation of Probabilistic Behavior
Table 2: Research Reagent Solutions for Probabilistic Behavioral Modeling
| Research Reagent | Function/Application | Specifications/Alternatives |
|---|---|---|
| Intertemporal Choice Task (ICT) | Presents series of choices between immediate smaller and delayed larger rewards to measure delay discounting [23] | Standardized task parameters: reward amounts, delay intervals, trial counts |
| Latent Variable Models | Probabilistically links behavioral observations to underlying cognitive processes using generative equations [23] | Various model architectures: exponential, hyperbolic discounting functions; softmax choice rules |
| Parameter Estimation Algorithms | Calibrates model parameters from individual choice sequences using maximum likelihood or Bayesian methods [23] | Optimization techniques: Markov Chain Monte Carlo, gradient descent, expectation-maximization |
| Adaptive Design Optimization | Generates experimental trials designed to elicit specific behavioral probabilities based on individual model parameters [23] | Algorithmic approaches: mutual information maximization, entropy minimization |
Experimental Workflow for Delay Discounting Studies:
This protocol exemplifies the modern probabilistic approach to modeling cognitive processes, with specific application to delay discounting behavior [23].
Materials and Setup:
- Implement computerized Intertemporal Choice Task (ICT) with precise timing controls
- Configure trial parameters: immediate reward (e.g., $10-$100), delayed reward (e.g., $20-$100), delay intervals (e.g., 1 day-1 year)
- Establish data collection system capturing choice responses and reaction times
Procedure:
- Run A (Model Inference):
- Present series of binary choice trials following standardized ICT protocol
- Collect minimum of 50-100 trials per participant to ensure reliable parameter estimation
- Record complete trial-by-trial data:
d = {(x_i, y_i), i = 1,2,...,T}wherex_iare predictor vectors of immediate/delayed options andy_iare observed choices [23]
Model Calibration:
- Estimate individual discounting parameters using maximum likelihood estimation
- Validate model fit through posterior predictive checks and residual analysis
- Compare alternative models using information criteria (AIC/BIC) or cross-validation
Run B (Model Application):
- Generate adaptive trials designed to induce specific discounting probabilities (0.1-0.9) using calibrated model
- Present second ICT with experimentally manipulated trial parameters based on model predictions
- Collect behavioral responses for validation of model predictions
Validation Analysis:
- Compare predicted versus observed choice probabilities across the induced probability range
- Calculate prediction error metrics (e.g., mean squared error, classification accuracy)
- Assess model validity through out-of-sample prediction performance [23]
Diagram 2: Experimental Workflow for Probabilistic Model Validation
Probabilistic Approaches to Model Verification and Validation
Framework for Probabilistic Model Validation
Modern probabilistic model validation represents a formal approach to assessing predictive accuracy while accounting for approximation error and uncertainty [24]. This framework is particularly relevant for computational models used in biomedical research, where both inherent variability (aleatory uncertainty) and limited knowledge (epistemic uncertainty) must be addressed.
The core validation procedure involves several key components:
Uncertainty Representation: Random quantities are represented using functional analytic approaches, particularly polynomial chaos expansions (PCEs), which permit the formulation of uncertainty assessment as a problem of approximation theory [24].
Parameter Calibration: Statistical procedures calibrate uncertain parameters from experimental or model-based measurements, using PCEs to represent inherent uncertainty of model parameters [24].
Hypothesis Testing: Simple hypothesis tests explore the validation of the computational model assumed for the physics (or biology) of the problem, comparing model predictions with experimental evidence [24].
Protocol: Probabilistic Model Checking for Biomedical Systems
Probabilistic model checking provides a formal verification approach for stochastic systems, with growing applications in biological modeling and healthcare systems [25].
Materials and Computational Resources:
- Probabilistic model checking software (PRISM, Storm, or Modest toolset)
- Model specification in appropriate formalism (DTMC, CTMC, MDP)
- Temporal logic properties (PCTL, CSL) encoding biological hypotheses
- High-performance computing resources for large state spaces
Procedure:
Model Formulation:
- Define system components and their probabilistic interactions
- Select appropriate modeling formalism based on system characteristics:
- Discrete-time Markov chains (DTMCs) for discrete probabilistic systems
- Continuous-time Markov chains (CTMCs) for systems with timing properties
- Markov decision processes (MDPs) for systems with nondeterminism and probability
Property Specification:
- Formalize biological hypotheses as temporal logic properties:
P≥0.95 [F≤100 "target_expression"](The probability that target expression level is eventually reached within 100 time units is at least 0.95)S≥0.98 ["steady_state"](The long-run probability of being in steady state is at least 0.98)
- Formalize biological hypotheses as temporal logic properties:
Model Checking Execution:
- Implement numerical algorithms for probability computation
- Handle state space explosion through:
- Symbolic methods using binary decision diagrams
- Statistical model checking for very large models
- Abstraction refinement techniques
Validation Metrics:
- Compare computation-observation agreement using statistical distance measures
- Implement validation metrics accounting for both aleatory and epistemic uncertainty
- Assess predictive capability through out-of-sample testing [24]
Emerging Trends and Future Directions
Integration of AI and Machine Learning
The life science analytics market is witnessing rapid integration of artificial intelligence and machine learning, particularly for drug discovery and development, personalized medicine, and disease management [21]. This trend is shifting the industry from initial AI hype toward "smart automation" that leverages the best automation approach—whether AI, rule-based, or other—to optimize efficiency and manage risk for each specific use case [22].
Key developments include:
AI-Augmented Medical Coding: Modified workflows where traditional rule-based automation handles most coding, with AI providing suggestions or automatic coding for remaining records [22]
Predictive Analytics Growth: The predictive segment is anticipated to grow with the highest CAGR in the life science analytics market, using statistical models and machine learning to forecast patient responses, identify clinical trial risks, and optimize market strategies [21]
Risk-Based Approaches: Regulatory encouragement of risk-based quality management (RBQM) is prompting sponsors to shift from traditional data collection to dynamic, analytical tasks focused on critical data points [22]
Advancing Model Validation Frameworks
Future developments in probabilistic model validation will need to address several challenging frontiers:
History-Dependent Models: Extension of validation frameworks to latent variable models with history dependence, where current behavior depends on previous states and choices [23]
Multi-Categorical Response Models: Development of validation approaches for response models with multiple categories beyond binary choice paradigms [23]
Uncertainty Quantification: Enhanced methods for characterizing both epistemic and aleatory uncertainties, particularly when dealing with limited experimental data [24]
The continued evolution of probabilistic analysis in biomedical research represents the modern instantiation of a centuries-long development toward more rigorous, quantitative assessment of medical evidence. From the "arithmetical observation" movement of the 18th century to contemporary AI-driven analytics, the fundamental goal remains consistent: to replace unfounded certainty with measured probability, thereby advancing both scientific understanding and clinical practice.
From Theory to Practice: Methodologies and Real-World Applications
Model-Informed Drug Development (MIDD) represents a paradigm shift in how pharmaceuticals are developed and evaluated. By leveraging quantitative modeling and simulation, MIDD provides a framework to integrate knowledge from diverse data sources, supporting more efficient and confident decision-making. These approaches allow researchers to extrapolate and interpolate information, enabling predictions of drug behavior in scenarios where direct clinical data may be limited or unavailable. The four methodological tools discussed in this article—Physiologically Based Pharmacokinetic (PBPK) modeling, Population Pharmacokinetic/Pharmacodynamic (PK/PD) modeling, Exposure-Response analysis, and Model-Based Meta-Analysis (MBMA)—form the cornerstone of modern MIDD. Within the context of model verification and validation, a probabilistic framework offers a rigorous methodology for quantifying uncertainty, assessing model credibility, and establishing the boundaries of reliable inference, thereby ensuring that model-based decisions are both scientifically sound and statistically justified [26] [27].
Physiologically Based Pharmacokinetic (PBPK) Modeling
PBPK modeling is a mathematical technique that predicts the absorption, distribution, metabolism, and excretion (ADME) of compounds by incorporating physiological, physicochemical, and biochemical parameters. Unlike traditional compartmental models that rely on empirical data fitting, PBPK models represent the body as a network of anatomically meaningful compartments, each corresponding to specific organs or tissues interconnected by the circulatory system [28] [26]. This physiological basis allows for a more mechanistic and realistic representation of drug disposition. The primary output of a PBPK simulation is a set of concentration-time profiles in plasma and various tissues, providing a comprehensive view of a drug's temporal behavior within the body [28].
Key Applications in Drug Development
Table 1: Key Applications of PBPK Modeling
| Application Area | Specific Use | Impact on Drug Development |
|---|---|---|
| Pediatric Drug Development | Extrapolating adult PK to children by incorporating age-dependent physiological changes [28]. | Reduces the need for extensive clinical trials in pediatric populations, addressing ethical and practical challenges. |
| Drug-Drug Interaction (DDI) Assessment | Predicting the potential for metabolic interactions when drugs are co-administered [26] [29]. | Informs contraindications and dosing recommendations, enhancing patient safety. |
| Formulation Optimization | Simulating absorption for different formulations (e.g., immediate vs. extended release) [26]. | Guides the selection of optimal formulation properties prior to costly manufacturing. |
| Special Population Dosing | Predicting PK in patients with organ impairment (e.g., hepatic or renal dysfunction) by adjusting corresponding physiological parameters [28] [29]. | Supports dose adjustment and labeling for subpopulations. |
| First-in-Human Dose Selection | Predicting safe and efficacious starting doses from preclinical data [29]. | De-risks early clinical trials and helps establish a rational starting point for dosing. |
Experimental Protocol for PBPK Model Development
The development of a whole-body PBPK model follows a systematic, stepwise protocol.
- Model Structure Specification: Define the model's anatomical representation. This involves selecting which organs and tissues to include as separate compartments based on the drug's properties and the model's purpose. Tissues with similar properties can be "lumped" together to simplify the model without sacrificing critical functionality [28].
- Tissue Model Selection: For each tissue compartment, determine the appropriate sub-model. The most common is the perfusion rate-limited (well-stirred) model, which assumes rapid equilibrium between blood and tissue. For drugs facing significant diffusion barriers (e.g., across the blood-brain barrier), a more complex permeability rate-limited model is required [28].
- Model Equation Formulation: Construct mass-balance differential equations for each compartment based on the law of mass action. These equations mathematically describe the rate of change of drug amount in each tissue as a function of blood flow, partitioning, and clearance processes [28].
- Model Parameterization: Populate the model with two distinct types of parameters:
- Physiological Parameters: Obtain values for organ volumes, blood flow rates, and other system-specific data from the scientific literature (e.g., International Commission on Radiological Protection (ICRP) for human values) [28].
- Compound-Specific Parameters: Incorporate drug-specific properties, which can be obtained from in vitro experiments or predicted using established algorithms. These include:
- Tissue-to-plasma partition coefficients (KpT), often predicted using algorithms by Poulin and Theil, or Rodgers and Rowland [28].
- Physicochemical properties like lipophilicity (LogP), pKa, and plasma protein binding (fu) [28].
- Clearance parameters, derived from in vitro metabolism data or in vivo studies [28].
- Model Verification and Validation (V&V): Execute the model and compare its predictions against observed in vivo data. This iterative process is crucial for establishing model credibility. A probabilistic V&V approach would involve assessing the uncertainty and sensitivity of key parameters, quantifying the probability that the model's predictions fall within an acceptable range of the observed data, and establishing a validation domain [28] [29].
Figure 1: PBPK Model Development and Probabilistic V&V Workflow. The process is iterative, with validation outcomes informing model refinement.
Research Reagent Solutions for PBPK
Table 2: Essential Research Reagents and Tools for PBPK Modeling
| Reagent/Tool Category | Specific Examples | Function in PBPK Workflow |
|---|---|---|
| In Vitro Assay Kits | Human liver microsomes (HLM); recombinant CYP enzymes; Caco-2 cell assays [28] [29]. | Generate compound-specific parameters for metabolism (e.g., CLint) and permeability. |
| Analytical Standards | Stable isotope-labeled drug analogs; certified reference standards. | Enable precise quantification of drug and metabolite concentrations in complex matrices for model validation. |
| Software Platforms | "Ready-to-use" platforms (e.g., GastroPlus, Simcyp); customizable modeling environments (e.g., MATLAB, R) [30]. | Provide the computational infrastructure for building, simulating, and validating PBPK models. |
| Physiological Databases | ICRP data; Brown et al. species-specific data [28]. | Source of reliable, literature-derived physiological parameters for model parameterization. |
| Partition Coefficient Predictors | Poulin & Theil; Rodgers & Rowland algorithms [28]. | In silico tools for estimating tissue:plasma partition coefficients from chemical structure and in vitro data. |
Population Pharmacokinetic and Pharmacodynamic (popPK/PD) Modeling
Population PK (popPK) is the study of the sources and correlates of variability in drug concentrations among individuals from a target patient population receiving therapeutic doses [31]. It uses nonlinear mixed-effects (NLME) models to analyze data from all individuals simultaneously. The "mixed-effects" terminology refers to the model's parameterization: fixed effects describe the typical parameter values (e.g., typical clearance) for the population, while random effects quantify the unexplained variability of these parameters between individuals (between-subject variability, BSV) and between occasions (between-occasion variability, BOV) [32]. popPD models then link these PK parameters to pharmacodynamic (PD) endpoints, describing how the drug's effect changes over time [33].
Key Applications in Drug Development
- Characterizing Variability: PopPK models identify and quantify the degree of inter-individual variability in PK parameters, which is essential for understanding the potential for variable exposure and response [31].
- Covariate Analysis: These models are used to identify patient-specific factors (covariates)—such as body weight, renal function, age, or genetics—that explain a portion of the random variability. This allows for dose individualization for specific subpopulations [32] [31].
- Sparse Data Analysis: A key strength of popPK is its ability to analyze data from late-phase trials where only a few samples (sparse data) are collected from each patient, making it ideal for studying PK in real-world clinical settings [32] [31].
- Clinical Trial Simulations: PopPK/PD models are used to simulate virtual clinical trials under different dosing regimens and patient demographics to optimize the design of future studies and assess the probability of technical success [31].
Experimental Protocol for PopPK Model Development
- Data Assembly and Curation: Compile a high-quality dataset containing drug concentration-time data, dosing records, and patient covariate information. Scrutinize data for errors and handle censored data (e.g., concentrations below the limit of quantification, BLOQ) appropriately, avoiding simplistic methods like imputing with zero or LLOQ/2 [32].
- Structural Model Development: Select a mathematical model that describes the typical concentration-time course. This is typically a mammillary compartmental model (e.g., one-, two-, or three-compartment) parameterized in terms of volumes (V) and clearances (CL) [32].
- Statistical Model Development: Define the models for random variability. This includes specifying the variance models for BSV (e.g., log-normal distributions for PK parameters), BOV, and residual unexplained variability (RUV), which accounts for measurement error and model misspecification [32].
- Covariate Model Building: Systematically test and incorporate relationships between patient covariates and PK parameters. For example, evaluate if creatinine clearance is a significant covariate on drug clearance. This is often done using a stepwise procedure guided by the Likelihood Ratio Test (LRT) on the difference in the Objective Function Value (OFV) [32].
- Model Evaluation: Assess the final model's performance using diagnostic plots (e.g., observed vs. population-predicted concentrations, conditional weighted residuals vs. time) and numerical methods (e.g., bootstrap, visual predictive check) [32].
- Probabilistic Model Validation: From a probabilistic V&V perspective, the model's predictive performance is quantified. This involves using techniques like the visual predictive check (VPC) to assess whether the variability and distribution of observed data fall within the prediction intervals of simulations from the final model, providing a probabilistic measure of model adequacy [32].
Figure 2: Population PK Model Development Workflow. The process emphasizes iterative evaluation and refinement to achieve a model that adequately describes population characteristics and variability.
Exposure-Response (E-R) Analysis
Exposure-Response (E-R) analysis is a critical component of popPK/PD that establishes the quantitative relationship between drug exposure (e.g., AUC, Cmax, Ctrough) and a pharmacodynamic outcome, which can be a measure of efficacy (e.g., change in a biomarker, clinical score) or safety (e.g., probability of an adverse event) [31]. The primary goal is to identify the exposure range that maximizes the therapeutic benefit while minimizing the risk of adverse effects, thereby defining the optimal dosing strategy.
Key Applications and Methodologies
- Dose Selection: E-R analysis is the foundation for selecting the recommended dose and dosing regimen for Phase 3 trials and eventual drug labeling [31].
- Benefit-Risk Assessment: It provides a quantitative framework for comparing the E-R relationships for efficacy and safety endpoints, enabling a holistic benefit-risk assessment [31].
- Concentration-QT (C-QT) Analysis: E-R modeling is used to characterize the effect of a drug on the heart's QT interval as a function of concentration. A well-developed C-QT model can sometimes serve as an alternative to a dedicated Thorough QT (TQT) study [31].
- Modeling Approaches: E-R relationships can be described by a variety of models, including direct-effect models (where effect is an immediate function of plasma concentration), indirect-effect models (where effect lags behind concentration), and more complex models for delayed effects, such as the transit compartment model or indirect response models.
Model-Based Meta-Analysis (MBMA)
Model-Based Meta-Analysis (MBMA) is a quantitative approach that integrates summary-level data from multiple published clinical trials, and potentially internal data, using pharmacological models to inform drug development decisions [34] [27]. Unlike traditional pairwise or network meta-analysis (NMA), which typically only use data from a single time point (e.g., the primary endpoint), MBMA incorporates longitudinal time-course data and dose-response relationships, providing a more dynamic and mechanistic view of the competitive landscape [34].
Key Applications in Drug Development
Table 3: Key Applications of Model-Based Meta-Analysis
| Application Area | Specific Use | Impact on Drug Development |
|---|---|---|
| Competitive Benchmarking | Comparing the efficacy and safety of an investigational drug against established standard-of-care treatments, even in the absence of head-to-head trials [34] [27]. | Informs target product profile (TPP) and differentiation strategy. |
| Optimal Dose Selection | Determining the dose and regimen for an internal molecule that is predicted to provide a competitive efficacy-safety profile based on external data [34]. | Increases confidence in Phase 3 dose selection. |
| Clinical Trial Optimization | Informing trial design by predicting placebo response, standard-of-care effect, and variability based on historical data [27]. | Improves trial power and efficiency; aids in go/no-go decisions. |
| Synthetic Control Arms | Creating model-based historical control arms for single-arm trials, providing a context for interpreting the results of the investigational treatment [27]. | Can reduce the need for concurrent placebo groups, accelerating development. |
| Market Access & Licensing | Evaluating the comparative value of an asset for business development and supporting reimbursement discussions with health technology assessment (HTA) bodies [27]. | Supports commercial strategy and in-licensing decisions. |
Experimental Protocol for MBMA
- Systematic Literature Review: Conduct a comprehensive and disciplined search of clinical literature in the therapeutic area of interest, following guidelines such as those in the Cochrane Handbook [34].
- Data Curation and Database Creation: Extract and curate aggregated data from the selected publications. This includes trial design features (doses, duration), patient baseline characteristics, and longitudinal efficacy/safety outcomes. Sophisticated databases like Certara's CODEX are used for this purpose [27].
- Base Model Development: Establish a model for the natural disease progression and placebo response over time. This serves as the foundation for evaluating drug effects [34].
- Drug Model Development: Incorporate the effect of active treatments. A common approach is to use an Emax model to describe the dose-response relationship, where the maximal effect (Emax) can be shared among drugs with the same mechanism of action (MOA) [34]. The model also accounts for the time-course of drug action, estimating parameters like the time to achieve 50% of the maximal effect (ET50) [34].
- Model Validation and Cross-Validation: Rigorously check model predictions against observed data from the literature and any held-out internal studies. This step is critical for assessing the model's predictive performance and identifying any systematic biases [34].
- Probabilistic Inference and Forecasting: Use the qualified MBMA to simulate clinical outcomes for new scenarios, such as the predicted performance of a novel drug candidate. The probabilistic framework involves quantifying uncertainty in all model parameters and propagating this uncertainty through simulations, resulting in predictions that are expressed as probability distributions (e.g., probability of success, probability of achieving a target differentiation) [34] [27].
Figure 3: Model-Based Meta-Analysis Workflow. The process transforms aggregated literature data into a dynamic predictive model for strategic decision-making.
Probabilistic model checking is a formal verification technique for analyzing stochastic systems against specifications expressed in temporal logic. This approach algorithmically checks whether a model of the system satisfies properties specified in temporal logic, enabling rigorous assessment of correctness, reliability, performance, and safety for systems incorporating random behavior [25]. The technique has evolved significantly from its origins in the 1980s, where early algorithms focused on verifying randomized algorithms, particularly in concurrent systems [25]. Today, probabilistic model checking supports a diverse set of modeling formalisms including Discrete-Time Markov Chains (DTMCs), Markov Decision Processes (MDPs), and Continuous-Time Markov Chains (CTMCs), with extensions to handle real-time constraints, costs, rewards, and partial observability [25].
The core temporal logics used in probabilistic model checking include Probabilistic Computation Tree Logic (PCTL) for DTMCs and MDPs, and Continuous Stochastic Logic (CSL) for CTMCs [25]. These logics enable specification of quantitative properties such as "the probability of system failure occurring is at most 0.01" or "the expected energy consumption before task completion is below 50 joules" [25] [35]. The integration of costs and rewards has further expanded the range of analyzable properties to include power consumption, resource utilization, and other quantitative system characteristics [25].
Foundational Concepts and Methodologies
Temporal Logic Foundations
Temporal logic provides the formal language for expressing system properties over time, forming the specification basis for model checking. Two primary types of temporal logic are employed:
Linear Temporal Logic (LTL) specifies properties along single execution paths, with formulas evaluated over sequences of states [36] [37]. Key LTL operators include:
- ○φ (Next): φ holds in the next state
- □φ (Always): φ holds in all future states
- ◇φ (Eventually): φ holds in some future state
- φ₁ 𝓤 φ₂ (Until): φ₁ holds until φ₂ becomes true
Computation Tree Logic (CTL) specifies properties over tree-like structures of possible futures, with path quantifiers (A for all paths, E for there exists a path) combined with temporal operators [36]. CTL enables reasoning about multiple possible futures simultaneously, making it suitable for nondeterministic systems.
Model checking involves a systematic state-space search to verify if a system model satisfies temporal logic specifications, with violations producing counterexamples that illustrate requirement breaches [37].
Probabilistic Modeling Formalisms
Different probabilistic modeling formalisms capture various aspects of system behavior:
Table 1: Probabilistic Modeling Formalisms and Their Applications
| Formalism | Key Characteristics | Representative Applications |
|---|---|---|
| Discrete-Time Markov Chains (DTMCs) | Discrete states, probabilistic transitions without nondeterminism | Randomized algorithms, communication protocols [25] |
| Markov Decision Processes (MDPs) | Combines probabilistic transitions with nondeterministic choices | Controller synthesis, security protocols, planning [25] |
| Continuous-Time Markov Chains (CTMCs) | Models continuous timing of events with exponential distributions | Performance evaluation, reliability analysis, queueing systems [25] |
| Probabilistic Timed Automata (PTA) | Extends MDPs with real-time clocks and timing constraints | Wireless communication protocols, scheduling problems [35] |
Application Domains and Case Studies
Communications and Network Protocols
Probabilistic model checking has been extensively applied to verify communication protocols, particularly those employing randomization for symmetry breaking or collision avoidance. The Ethernet protocol with its exponential back-off scheme represents a classic case where probabilistic verification ensures reliable operation under uncertain message delays and losses [25]. Notable successes include:
- Bluetooth Protocol Verification: Analysis of DTMC models with over 10¹⁰ states to evaluate differences between protocol variants and underlying model assumptions [25]
- Wireless Sensor Networks: Verification of reliability and timeliness properties in resource-constrained environments where hardware limitations magnify the importance of unreliability analysis [25]
- Token-Passing Schemes: Comparison of wireless token-passing approaches for safety-critical systems using probabilistic timed automata [25]
These applications demonstrate how probabilistic model checking can quantify reliability and performance metrics for network designs, often employing CSL for properties such as "the probability of server response exceeding 1 second is at most 0.02" or "long-run network availability is at least 98%" [25].
Computer Security
Security analysis represents another significant application area, where MDPs effectively model the interplay between adversarial actions (nondeterminism) and system randomization (probability) [25]. Security protocols frequently incorporate randomness for key generation, session identifier creation, and prevention of attacks like buffer overflows or DNS cache poisoning [25]. Probabilistic model checking enables formal verification of security properties despite potential adversarial behavior, providing strong guarantees about system resilience.
Pharmaceutical Development and Drug Discovery
In pharmaceutical development, the Probability of Pharmacological Success (PoPS) framework adapts probabilistic assessment to evaluate drug candidates during early development stages [38]. PoPS represents the probability that most patients achieve adequate pharmacology for the intended indication while minimizing subjects exposed to safety risks [38]. This application demonstrates how probabilistic formal methods extend beyond traditional computing systems to complex biological domains.
The PoPS calculation incorporates multiple uncertainty sources through a structured methodology:
- Define Success Criteria: Establish thresholds for adequate pharmacology and acceptable safety risk based on scientific evidence, which may include genetic evidence, in vivo/ex vivo experiments, or clinical data from similar drugs [38]
- Develop PK/PD Models: Create pharmacokinetic/pharmacodynamic models based on in vitro, animal, or early human data, capturing between-subject variability and parameter uncertainty [38]
- Translate to Patient Population: Adapt models to target patient populations, accounting for translation uncertainty [38]
- Simulate Virtual Populations: Generate multiple virtual patient populations using Monte Carlo approaches [38]
- Compute Success Probability: Calculate PoPS as the proportion of virtual populations meeting success criteria [38]
This methodology integrates multi-source data, identifies knowledge gaps, and enforces transparency in assumptions, supporting more rigorous dose prediction and candidate selection decisions [38].
Experimental Protocols and Workflows
Protocol for Probabilistic Model Checking of System Designs
This protocol outlines the general procedure for applying probabilistic model checking to system verification, adaptable to various domains including communications, security, and biological systems.
Table 2: Probabilistic Model Checking Protocol
| Step | Description | Tools/Techniques |
|---|---|---|
| 1. System Modeling | Abstract system components as states and transitions in appropriate formalism (DTMC, MDP, CTMC) | PRISM, Storm, Modest toolset [25] |
| 2. Property Specification | Formalize requirements using temporal logic (PCTL, CSL) with probabilistic and reward operators | PCTL for DTMCs/MDPs, CSL for CTMCs [25] |
| 3. Model Construction | Build state transition representation, applying reduction techniques for large state spaces | Symbolic methods using binary decision diagrams [25] |
| 4. Property Verification | Algorithmically check specified properties against the model | Probabilistic model checking algorithms [25] |
| 5. Result Analysis | Interpret quantitative results, generate counterexamples for violated properties | Visualization tools, counterexample generation [36] |
Figure 1: Probabilistic Model Checking Workflow
Protocol for Pharmaceutical PoPS Assessment
This specialized protocol details the Probability of Pharmacological Success assessment for early-stage drug development decisions, based on methodologies successfully implemented in pharmaceutical companies [38].
Table 3: PoPS Assessment Protocol for Drug Development
| Step | Description | Key Considerations |
|---|---|---|
| 1. Define Success Criteria | Establish thresholds for adequate pharmacology and acceptable safety | Criteria should reflect levels expected to produce clinical efficacy, not intuition [38] |
| 2. Develop PK/PD Models | Create exposure-response models for pharmacology and safety endpoints | Incorporate between-subject variability and parameter estimation uncertainty [38] |
| 3. Translate to Humans | Adapt models from preclinical data to patient populations | Account for translation uncertainty between species or experimental conditions [38] |
| 4. Simulate Virtual Populations | Generate virtual patient data using Monte Carlo methods | Typical virtual population size: N=1000 patients; parameter simulations: M=500 [38] |
| 5. Compute PoPS | Calculate probability of meeting success criteria across simulations | PoPS = M'/M, where M' is number of successful virtual populations [38] |
Figure 2: Pharmaceutical PoPS Assessment Workflow
Research Reagent Solutions and Tools
The effective application of probabilistic model checking requires specialized software tools and modeling frameworks. The following table catalogs essential resources for researchers implementing these techniques.
Table 4: Essential Research Tools for Probabilistic Model Checking
| Tool/Resource | Type | Primary Function | Application Context |
|---|---|---|---|
| PRISM [25] | Software Tool | General-purpose probabilistic model checker | Multiple domains: randomized algorithms, security, biological systems |
| Storm [25] | Software Tool | High-performance probabilistic model checking | Analysis of large, complex models requiring efficient computation |
| Modest Toolset [25] | Software Tool Suite | Modeling and analysis of stochastic systems | Systems with hard and soft timing constraints |
| UPPAAL [35] | Software Tool | Verification of real-time systems | Probabilistic timed automata, scheduling problems |
| NuSMV [36] | Software Tool | Symbolic model checker | CTL model checking, finite-state systems |
| SPIN [36] | Software Tool | Model checker for distributed systems | LTL model checking, concurrent software verification |
| PoPS Framework [38] | Methodology | Probability of pharmacological success assessment | Early-stage drug development decisions |
Advanced Applications and Future Directions
Probabilistic model checking continues to expand into new domains and incorporate advanced modeling capabilities. Emerging applications include:
- Partially Observable MDPs: Modeling autonomous systems with unreliable sensors or security protocols with secret keys [25]
- Stochastic Games: Reasoning about competitive or collaborative agents in probabilistic settings [25]
- Uncertain Markov Models: Capturing epistemic uncertainty from data-driven modeling [25]
- AI System Verification: Applying formal verification to neural networks and AI-based software systems [39]
The cross-fertilization between verification techniques is also advancing, with growing integration between verification, model checking, and abstract interpretation methods [40]. These hybrid approaches offer promising avenues for addressing the scalability challenges inherent in analyzing complex, real-world systems.
In pharmaceutical applications, PoPS methodologies are evolving to incorporate diverse data sources including real-world evidence and historical clinical trial data, improving the accuracy of drug development decision-making [41] [38]. As these techniques mature, they enable more rigorous assessment of benefit-risk ratios throughout the drug development pipeline, potentially reducing attrition rates and improving resource allocation.
The continued development of probabilistic model checking tools and methodologies ensures their expanding relevance across diverse domains, from traditional computer systems to emerging applications in healthcare, biological modeling, and artificial intelligence.
Implementing AI and Machine Learning for Enhanced Predictive Modeling
Application Note: Current AI Landscape in Drug Discovery and Development
The application of artificial intelligence (AI) and machine learning (ML) in drug discovery represents a paradigm shift, moving from labor-intensive, human-driven workflows to AI-powered discovery engines that compress timelines and expand investigational search spaces [42]. By 2025, AI has progressed from experimental curiosity to clinical utility, with AI-designed therapeutics now in human trials across diverse therapeutic areas [42]. These technologies are being applied across the entire drug development continuum, from initial target identification to clinical trial optimization [43] [44].
Leading AI-driven platforms have demonstrated remarkable capabilities in accelerating early-stage research. For instance, Insilico Medicine's generative-AI-designed idiopathic pulmonary fibrosis drug progressed from target discovery to Phase I trials in just 18 months, significantly faster than the typical 5-year timeline for traditional discovery and preclinical work [42]. Similarly, Exscientia reports in silico design cycles approximately 70% faster than industry standards, requiring 10 times fewer synthesized compounds [42]. These accelerations are made possible through several specialized AI approaches, as implemented by leading platforms detailed in Table 1.
Table 1: Leading AI-Driven Drug Discovery Platforms and Their Applications
| Platform/Company | Core AI Approach | Key Applications | Clinical Stage Achievements |
|---|---|---|---|
| Exscientia [42] | Generative Chemistry, Centaur Chemist | Integrated target-to-design pipeline, Automated design-make-test-learn cycle | First AI-designed drug (DSP-1181) in Phase I for OCD; Multiple clinical compounds for oncology and inflammation |
| Insilico Medicine [42] | Generative AI | Target discovery, De novo molecular design | Phase IIa results for TNIK inhibitor in idiopathic pulmonary fibrosis; Target-to-Phase I in 18 months |
| Recursion [42] | Phenomics-First Systems | High-content phenotypic screening, Biological data mining | Merged with Exscientia (2024) to integrate phenomics with generative chemistry |
| Schrödinger [42] | Physics-plus-ML Design | Molecular simulations, Physics-enabled drug design | TYK2 inhibitor (zasocitinib) advanced to Phase III trials |
| BenevolentAI [42] | Knowledge-Graph Repurposing | Target identification, Drug repurposing, Biomedical data integration | Multiple candidates in clinical stages for inflammatory and neurological diseases |
The proliferation of these platforms has led to substantial growth in AI-derived clinical candidates, with over 75 molecules reaching clinical stages by the end of 2024 [42]. This growth underscores the expanding role of AI in reshaping pharmacological research worldwide.
Application Note: AI for Molecular Design and Optimization
Generative AI for Molecular Design
Generative AI models have emerged as powerful tools for creating novel molecular structures targeting specific disease pathways. The BoltzGen model developed at MIT represents a significant advancement as the first model capable of generating novel protein binders ready to enter the drug discovery pipeline [45]. Unlike previous models limited to specific protein types or easy targets, BoltzGen implements three key innovations: (1) unification of protein design and structure prediction tasks while maintaining state-of-the-art performance; (2) built-in biophysical constraints informed by wet-lab collaborations to ensure generated proteins are functional and physically plausible; and (3) rigorous evaluation on challenging "undruggable" disease targets [45].
This approach moves beyond simple pattern matching to emulate underlying physical principles. As developer Hannes Stärk explains, "A general model does not only mean that we can address more tasks. Additionally, we obtain a better model for the individual task since emulating physics is learned by example, and with a more general training scheme, we provide more such examples containing generalizable physical patterns" [45]. The model was comprehensively validated on 26 diverse targets in eight independent wet labs across academia and industry, demonstrating its potential for breakthrough drug development [45].
Predictive Modeling in Pharmacokinetics
In pharmacokinetics (PK), AI-based models are challenging traditional gold-standard methods like nonlinear mixed-effects modeling (NONMEM). A 2025 comparative study evaluated five machine learning models, three deep learning models, and a neural ordinary differential equations (ODE) model on both simulated and real clinical datasets [46]. The results demonstrated that AI/ML models often outperform NONMEM, with variations in performance depending on model type and data characteristics [46].
Notably, neural ODE models showed particularly strong performance, providing both predictive accuracy and explainability with large datasets [46]. This is significant for model-informed drug development (MIDD), where understanding the biological basis of predictions is crucial for regulatory acceptance and clinical decision-making. The integration of these AI methodologies into pharmacometric workflows presents opportunities to enhance predictive performance and computational efficiency in optimizing drug dosing strategies [46].
Protocol: Probabilistic Verification and Validation of AI Models
Unified V&V Methodology for AI-Driven Predictive Models
The deployment of AI models in safety-critical drug development applications necessitates rigorous verification and validation (V&V) frameworks. A unified V&V methodology addresses this need by combining formal verification with statistical validation to provide robust safety guarantees [17]. This approach is particularly relevant for vision-based autonomous systems in laboratory automation and for validating AI models that operate under distribution shifts between training and deployment environments.
The methodology consists of three integrated steps:
Abstraction: Construct an interval Markov decision process (IMDP) abstraction that represents neural perception uncertainty with confidence intervals. This abstraction overapproximates the concrete system with a specified confidence level (e.g., with probability 1-α, the IMDP contains the behavior distribution of the true system) [17].
Verification: Apply probabilistic model checking to the constructed IMDP to verify system-level temporal properties. This step produces an upper bound β on the probability that a trajectory violates the safety property φ [17].
Validation: Validate the IMDP abstraction against new deployment environments by constructing a belief over the parameters of the new environment and computing the posterior probability (1-γ) that the new environment falls within the uncertainty bounds of the original IMDP [17].
This workflow results in a nested probabilistic guarantee: with confidence 1-γ, the system in the new environment satisfies the safety property φ with probability at least 1-β [17]. The following diagram illustrates this integrated methodology and the probabilistic relationships between its components.
Experimental Protocol for V&V of AI Models
Purpose: To empirically verify and validate the safety and performance of AI-based predictive models under domain shift conditions.
Materials and Reagents: Table 2: Research Reagent Solutions for AI Model V&V
| Item | Function | Example Implementation |
|---|---|---|
| Interval Markov Decision Process (IMDP) Framework | Provides formal abstraction for modeling uncertainty in perception and dynamics | IMDP abstraction ℳ_E with probability intervals [17] |
| Probabilistic Model Checker | Verifies temporal logic properties against probabilistic models | Tools like PRISM, Storm, or PAYNT [47] |
| Validation Dataset from New Environment | Quantifies model performance under domain shift | Dataset from E' with distribution different from training data E [17] |
| Bayesian Inference Tool | Computes posterior belief over model parameters | Probabilistic programming languages (e.g., Pyro, Stan) [17] |
| Formal Specification Library | Encodes safety requirements as verifiable properties | Temporal logic formulae (e.g., PCTL, LTL) defining safety constraints [17] |
Procedure:
Baseline Model Training:
- Train the target AI model (e.g., a convolutional neural network for cellular image analysis [43] or a protein binder generator [45]) on the original training dataset from environment E.
- Document baseline performance metrics (e.g., root mean squared error (RMSE), mean absolute error (MAE), coefficient of determination (R²) [46]) on a held-out test set from the same environment.
IMDP Abstraction Construction:
- From the trained model and training data, construct an IMDP abstraction ℳ_E that overapproximates the system behavior with confidence level α (typically α=0.05 for 95% confidence) [17].
- The IMDP should capture uncertainties in both the perception component (e.g., neural network predictions) and system dynamics.
Formal Verification:
- Encode the safety property φ as a temporal logic formula (e.g., "the system never enters an unsafe state" or "the system eventually reaches a target state with probability at least p").
- Use a probabilistic model checker to verify φ on the IMDP abstraction ℳ_E, obtaining the verification bound β [17].
- This yields the frequentist guarantee: with confidence 1-α, the original system M_E satisfies P(φ) ≥ 1-β.
Domain Shift Validation:
- Deploy the trained model in a new environment E' with different distribution characteristics (e.g., different cellular imaging conditions [43] or novel protein targets [45]).
- Collect a validation dataset from E' and compute the posterior probability 1-γ that the new environment's concrete model ME' is contained within the abstraction ℳE using Bayesian inference [17].
Nested Guarantee Calculation:
- Combine the results from verification and validation to establish the nested probabilistic guarantee: with confidence 1-γ, the system in environment E' satisfies the safety property φ with probability at least 1-β [17].
Troubleshooting:
- If the validation step yields a low posterior (1-γ < 0.95), consider refining the IMDP abstraction with data from the new environment or implementing domain adaptation techniques.
- If verification produces excessively conservative bounds (β close to 1), examine the IMDP construction for overly wide probability intervals and refine the uncertainty quantification.
Protocol: Implementation of Generative AI for Protein Binder Design
Experimental Workflow for Novel Protein Binder Generation
Purpose: To generate novel protein binders targeting specific disease-associated proteins using generative AI models.
Materials:
- Target protein structure (e.g., from AlphaFold DB or Protein Data Bank)
- BoltzGen or similar generative protein design model [45]
- Molecular dynamics simulation software (e.g., GROMACS, AMBER)
- Wet-lab facilities for synthesis and validation (e.g., phage display, surface plasmon resonance)
The following diagram outlines the integrated computational and experimental workflow for generating and validating AI-designed protein binders, highlighting the critical role of probabilistic verification at key stages.
Procedure:
Target Identification and Preparation:
- Select a therapeutic target protein, preferably from the "undruggable" category to demonstrate capability against challenging targets [45].
- Obtain or generate the 3D structure of the target protein through experimental methods (e.g., crystallography) or computational prediction (e.g., AlphaFold).
Generative AI Model Setup:
- Configure the BoltzGen model or equivalent generative protein design platform.
- Input the target protein structure and set appropriate constraints based on feedback from wet-lab collaborators to ensure physical plausibility and functionality [45].
Binder Generation and In Silico Validation:
- Execute the generative model to produce candidate protein binders.
- Screen generated binders using in silico methods including:
- Molecular docking simulations to predict binding affinity and pose
- Molecular dynamics simulations to assess binding stability
- Physicochemical property prediction to ensure drug-like characteristics
Probabilistic Verification:
- Apply formal verification methods to assess the reliability of the AI-generated designs.
- Use IMDP abstractions to model uncertainties in the binding predictions and compute confidence bounds on binding affinity estimates [17].
- This verification step provides a quantitative measure of confidence in the AI-generated designs before proceeding to costly synthesis and experimental validation.
Experimental Validation:
- Synthesize top-ranking candidate binders identified through computational screening.
- Perform in vitro binding assays (e.g., surface plasmon resonance, isothermal titration calorimetry) to measure binding affinity and kinetics.
- Conduct functional assays in cellular models to assess biological activity and therapeutic potential.
Model Refinement:
- Incorporate experimental results as feedback to refine the generative AI model.
- Iterate the design process to optimize binder properties based on experimental data.
Troubleshooting:
- If generated binders show poor stability in simulations, adjust the biophysical constraints in the generative model to enforce stricter stability criteria.
- If experimental binding affinities do not correlate with computational predictions, recalibrate the scoring functions using the experimental data.
Table 3: Key Computational Tools and Platforms for AI-Driven Predictive Modeling
| Tool Category | Specific Tools/Platforms | Function | Application Context |
|---|---|---|---|
| Generative AI Models | BoltzGen [45], Exscientia Centaur Chemist [42], Insilico Medicine Generative Platform [42] | De novo molecular design, Protein binder generation | Creating novel therapeutic candidates against hard-to-treat targets |
| Probabilistic Verification | IMDP Abstraction Tools [17], PRISM [47], Storm, PAYNT [47] | Formal verification of safety properties, Uncertainty quantification | Providing rigorous safety guarantees for AI models in critical applications |
| Pharmacometric Modeling | Neural ODEs [46], NONMEM [46], Machine Learning PK Models [46] | Population pharmacokinetic prediction, Drug dosing optimization | Predicting drug behavior in diverse patient populations |
| Data Analysis & Visualization | Bayesian Inference Tools [17], Probabilistic Programming Languages (Pyro, Stan) | Statistical validation, Posterior probability computation | Quantifying model validity under domain shift conditions |
| Specialized AI Platforms | Recursion Phenomics [42], Schrödinger Physics-ML [42], BenevolentAI Knowledge Graphs [42] | Target identification, Lead optimization, Drug repurposing | Multiple stages of drug discovery from target validation to candidate optimization |
The integration of Probabilistic Physiologically Based Pharmacokinetic/Pharmacodynamic (PBPK/PD) modeling represents a paradigm shift in modern drug discovery and development. This approach combines mechanistic mathematical modeling with quantitative uncertainty analysis to predict drug behavior and effects across diverse populations, thereby addressing critical challenges in compound selection, dose optimization, and clinical translation [48] [49]. The probabilistic framework explicitly accounts for physiological variability and parameter uncertainty, moving beyond deterministic simulations to provide a more comprehensive risk-assessment framework for decision-making.
This case study illustrates the application of probabilistic PBPK/PD modeling within integrated drug discovery, framed within the broader context of model verification and validation research. We demonstrate how this approach enables quantitative prediction of interindividual variability arising from genetic polymorphisms, life-stage changes, and disease states, ultimately supporting the development of personalized dosing strategies and de-risked clinical development paths [48].
Background and Significance
Evolution of Model-Informed Drug Development
Model-Informed Drug Development (MIDD) has emerged as an essential framework that leverages quantitative methods to inform drug development decisions and regulatory evaluations [49]. The history of MIDD has significantly benefited from collaborative efforts between pharmaceutical sectors, regulatory agencies, and academic institutions. Recent harmonization initiatives, such as the ICH M15 guidance, promise to improve consistency in applying MIDD across global regulatory jurisdictions [49].
Traditional PK/PD modeling often relies on Hill-based equations (e.g., sigmoidal Emax model) to link drug concentration and effect, assuming rapid equilibrium between drug and target [50]. However, these approaches have limitations, particularly for drugs with slow target dissociation kinetics or those operating through non-equilibrium mechanisms. Approximately 80% of FDA-approved drugs between 2001-2004 operated through such non-equilibrium mechanisms, highlighting the importance of incorporating binding kinetics into predictive models [50].
The Probabilistic PBPK/PD Approach
Probabilistic PBPK/PD modeling extends conventional approaches through:
- Mechanistic integration of physiological processes, drug-target interaction kinetics, and system-specific parameters
- Quantitative uncertainty propagation using Monte Carlo simulations and related techniques
- Population variability representation through parameter distributions rather than point estimates
- Sensitivity analysis to identify critical parameters driving variability in outcomes
This approach is particularly valuable for predicting interindividual variability in special populations where clinical testing raises ethical concerns, including pregnant women, pediatric and geriatric patients, and individuals with organ impairments [48].
Methods and Experimental Protocols
Probabilistic PBPK Model Development
Objective: To develop a probabilistic PBPK model that predicts drug concentration-time profiles in diverse human populations, accounting for physiological variability and uncertainty in parameter estimates.
Protocol:
System Characterization
- Define the anatomical structure (organs and tissues connected by circulatory system)
- Specify physiological parameters (tissue volumes, blood flow rates) as probability distributions based on population data [48]
- Identify compound-specific parameters (logP, pKa, blood-to-plasma ratio) with associated uncertainty
Model Parameterization
- Gather in vitro absorption data (e.g., Caco-2 permeability, solubility)
- Obtain distribution parameters (e.g., tissue-to-plasma partition coefficients using in vitro or in silico methods)
- Characterize metabolism and excretion (e.g., CLint from microsomal stability studies, transporter kinetics)
- Define variability distributions for all parameters using literature data or experimental replicates
Model Implementation
- Code the model structure using differential equations to represent mass balance
- Implement Monte Carlo simulation framework for probabilistic analysis
- Validate model against in vivo preclinical PK data (rat, dog, or monkey)
Model Verification
- Conduct sensitivity analysis to identify influential parameters
- Perform qualitative verification against known physiological principles
- Execute quantitative verification comparing deterministic simulation to established solutions
Table 1: Key Physiological Parameters for PBPK Modeling with Associated Variability
| Parameter | Mean Value | Distribution Type | CV% | Source |
|---|---|---|---|---|
| Cardiac Output (L/h) | 16.2 | Normal | 15 | [48] |
| Liver Volume (L) | 1.4 | Lognormal | 20 | [48] |
| CYP2D6 Abundance (pmol/mg) | 5.8 | Bimodal | 40* | [48] |
| GFR (mL/min) | 117 | Normal | 18 | [48] |
| Intestinal Transit Time (h) | 2.1 | Weibull | 25 | - |
*Highly polymorphic enzymes show greater variability
Binding Kinetics-Based PD Model
Objective: To develop a mechanistic PD model that explicitly incorporates the kinetics of drug-target interactions, replacing traditional Hill-based models.
Protocol:
Target Engagement Characterization
- Determine binding kinetics (kon, koff) using surface plasmon resonance (SPR) or similar techniques
- Calculate residence time (τ = 1/koff)
- Measure IC50/EC50 under pre-equilibrium and equilibrium conditions
- Establish target vulnerability function linking occupancy to effect [50]
Cellular Effect Modeling
- Develop cell system model incorporating:
- Drug transport (passive/facilitated)
- Intracellular binding kinetics
- Downstream signaling pathways
- Physiological effect readout
- Parameterize using in vitro time-course data with multiple concentrations
- Develop cell system model incorporating:
Integrated PBPK/PD Implementation
- Link PBPK output to PD model input
- Implement target turnover dynamics for relevant targets
- Incorporate disease progression components where applicable
- Validate against preclinical efficacy models
Probabilistic Model Verification and Validation Framework
Objective: To establish a comprehensive framework for verifying and validating probabilistic PBPK/PD models within the context of regulatory decision-making.
Protocol:
Verification Phase
- Code verification: Ensure computational implementation accurately represents mathematical model
- Numerical verification: Confirm solution accuracy and stability across parameter space
- Unit consistency check: Verify dimensional homogeneity throughout model
Validation Phase
- Internal validation: Compare model predictions to data used for model development
- Calculate goodness-of-fit metrics (R², AIC, BIC)
- Perform visual predictive checks
- Execute residual analysis
- External validation: Test model against independent datasets not used in development
- Assess predictive performance using metrics like fold error, RMSE
- Conduct population predictive checks comparing simulated vs. observed variability
- Cross-validation: Implement k-fold or leave-one-out approaches for limited datasets
- Internal validation: Compare model predictions to data used for model development
Uncertainty Quantification
- Perform global sensitivity analysis (e.g., Sobol method, Morris screening)
- Execute uncertainty propagation through Monte Carlo simulation
- Generate prediction intervals with confidence levels
Context of Use Assessment
- Define model purpose and acceptance criteria based on context of use [49]
- Document model limitations and domain of applicability
- Establish credibility evidence based on risk associated with decision
Results and Application
Implementation of Probabilistic PBPK/PD Modeling
The implementation workflow for probabilistic PBPK/PD modeling involves multiple interconnected components, as illustrated below:
Diagram 1: Probabilistic PBPK/PD Modeling Workflow
Application in Special Population Dosing
A key application of probabilistic PBPK/PD modeling is predicting appropriate dosing regimens for special populations where clinical trials are ethically challenging or impractical. The following table summarizes quantitative findings for metabolic enzyme polymorphisms across biogeographical groups:
Table 2: CYP2D6 Phenotype Frequencies Across Populations [48]
| Population Group | Ultrarapid Metabolizer (%) | Normal Metabolizer (%) | Intermediate Metabolizer (%) | Poor Metabolizer (%) |
|---|---|---|---|---|
| European | 2 | 49 | 38 | 7 |
| East Asian | 1 | 53 | 38 | 1 |
| Sub-Saharan African | 4 | 46 | 38 | 2 |
| Latino | 4 | 60 | 29 | 3 |
| Central/South Asian | 2 | 58 | 28 | 2 |
| Near Eastern | 7 | 57 | 30 | 2 |
These population-specific polymorphism data can be incorporated into probabilistic PBPK models to simulate exposure differences and optimize dosing strategies for different ethnic groups, demonstrating the value of this approach in personalized medicine.
Model Verification and Validation Results
The verification and validation process for probabilistic models follows a rigorous pathway:
Diagram 2: Model Verification and Validation Pathway
Application of this framework to a case study involving LpxC inhibitors for antibacterial development demonstrated superior performance compared to traditional rapid-equilibrium models. The kinetics-driven model successfully predicted in vivo efficacy where traditional approaches significantly underestimated the required dose, highlighting the importance of incorporating drug-target residence time [50].
The Scientist's Toolkit
Research Reagent Solutions
Table 3: Essential Research Reagents for Probabilistic PBPK/PD Modeling
| Reagent/Category | Function/Application | Example Products |
|---|---|---|
| Human Liver Microsomes | CYP450 metabolism studies | Xenotech, Corning |
| Recombinant CYP Enzymes | Reaction phenotyping | Supersomes (Corning) |
| Transfected Cell Systems | Transporter activity assessment | Solvo Transporter Assays |
| SPR/BLI Platforms | Binding kinetics measurement | Biacore, Octet |
| Primary Hepatocytes | Hepatic clearance prediction | BioIVT, Lonza |
| Tissue Homogenates | Tissue partition coefficient determination | BioIVT, XenoTech |
| Biomarker Assays | Target engagement verification | MSD, Luminex |
| Probabilistic Modeling Software | Uncertainty quantification | R, Python, MATLAB |
Computational Tools and Platforms
Successful implementation of probabilistic PBPK/PD modeling requires specialized computational tools:
- PBPK Platforms: GastroPlus, Simcyp, PK-Sim
- Probabilistic Analysis: R with mrgsolve/PopED, Python with PyMC3/Stan
- Model Verification: Feature-based testing frameworks, unit test implementations
- Sensitivity Analysis: SALib, SimBiology
- Visualization: ggplot2, Matplotlib, Plotly
Discussion and Future Perspectives
The case study demonstrates that probabilistic PBPK/PD modeling provides a powerful framework for integrating diverse data sources and quantifying uncertainty in drug development predictions. By explicitly incorporating population variability and parameter uncertainty, this approach enables more informed decision-making throughout the drug development pipeline.
The verification and validation framework presented establishes rigorous standards for assessing model credibility within specific contexts of use. This is particularly important as regulatory agencies increasingly accept modeling and simulation evidence in support of drug approvals [49]. Future developments in this field will likely focus on:
- AI/ML integration for parameter estimation and model refinement [49]
- Enhanced biomarker integration for improved target engagement assessment [51]
- High-performance computing applications for complex population simulations
- Standardized verification protocols across the pharmaceutical industry
- Expanded application to novel therapeutic modalities
The probabilistic approach to PBPK/PD modeling represents a significant advancement in model-informed drug development, with potential to reduce late-stage failures, optimize resource allocation, and ultimately accelerate the delivery of effective therapies to patients.
The 505(b)(2) regulatory pathway, established under the Hatch-Waxman Amendments, represents a strategic hybrid approach to new drug approval that balances innovation with efficiency [52]. This pathway allows sponsors to leverage existing data from previously approved drugs or published literature, exempting them from repeating all original development work [52]. For drug development professionals, this pathway significantly reduces time and capital investment compared to the traditional 505(b)(1) route while creating differentiated products that escape pure generic competition [52]. This application note details how a probabilistic approach to model verification and validation can be systematically integrated into 505(b)(2) development programs, enhancing decision-making confidence and regulatory success.
Regulatory Pathway Comparison
The 505(b)(2) pathway occupies a strategic middle ground between innovative new chemical entities and generic copies, enabling modifications and improvements to existing therapies [52]. Understanding its distinction from other pathways is fundamental to strategic planning.
Table 1: Comparison of FDA Drug Approval Pathways [52]
| Feature | 505(b)(1) (Full NDA) | 505(b)(2) (Hybrid NDA) | 505(j) (ANDA – Generic) |
|---|---|---|---|
| Purpose | Approval for a completely new drug product/NME | Approval for modified versions of previously approved drugs | Approval for generic versions of Reference Listed Drugs (RLDs) |
| Data Reliance | Full preclinical & clinical data generated by applicant | Relies partly on existing data (literature, FDA findings) + new bridging studies | Focus on bioequivalence to RLD, no new clinical trials |
| Innovation Level | Significant innovation (new molecule/mechanism) | Innovation in formulation, dosage, route, indication, combination, etc. | Little to no new innovation (a "copy") |
| Development Time | Longest (up to 15 years) | Moderate (faster than 505(b)(1)) | Fastest |
| Development Cost | Highest (billions) | Moderate (more than 505(j), less than 505(b)(1)) | Lowest |
| Market Exclusivity | 5 years (NCE) + others (e.g., pediatric) | 3-7 years (e.g., 3-year "other" exclusivity, 7-year orphan) | 180-day first-filer exclusivity |
The core principle of 505(b)(2) is the leveraged use of existing data not generated by the applicant, which significantly reduces the need for duplicative, resource-intensive studies [52] [53]. The clinical pharmacology program under 505(b)(2) is typically more streamlined than for 505(b)(1), often relying on successful scientific bridging to the Listed Drug (LD) for aspects like Mechanism of Action (MOA), ADME properties, and the impact of intrinsic/extrinsic factors [53].
Probabilistic Framework for 505(b)(2) Development
A probabilistic approach provides a quantitative foundation for decision-making throughout the 505(b)(2) development lifecycle. This involves creating predictive models and formally verifying their correctness to de-risk development.
Core Principles and Model Architecture
Probabilistic models quantify the likelihood of successful outcomes based on known inputs and historical data. In the context of 505(b)(2) development, this can be applied to predicting bioequivalence success, optimizing clinical trial parameters, and forecasting regulatory approval probabilities. The model architecture integrates data on drug properties, study design, and historical performance metrics to generate predictive outputs with confidence intervals [7] [54].
Formal verification of these models, using tools and methodologies such as the Event-B formal method and the Rodin platform, ensures algorithmic correctness and reliability through mathematical proof [7]. This process involves constructing proof obligations and generating automated or manual proofs to verify that the model's invariants hold under all specified conditions [7].
Application to Key Development Scenarios
- Predicting Bioequivalence (BE) Study Success: A probabilistic model can integrate factors such as in vitro-in vivo correlation (IVIVC) data, formulation variability, and prior BE success rates for similar chemical entities. The output is a probability distribution of achieving BE, guiding formulation optimization before costly clinical studies.
- Optimizing Clinical Pharmacology Programs: For a drug seeking approval for a new route of administration, a model can probabilistically weigh the need for specific studies (e.g., renal impairment, drug-drug interactions) based on the extent of systemic exposure change and available data from the reference drug [53]. Model-Informed Drug Development (MIDD) approaches are key enablers here [53].
- Quantifying Regulatory Risk: By analyzing the historical success rates of similar 505(b)(2) submissions (e.g., changes from immediate-release to extended-release), a model can assign a probability of approval, helping portfolio managers prioritize projects.
The following workflow diagram illustrates the integration of probabilistic modeling and verification within a 505(b)(2) development program.
Experimental Protocols
This section provides detailed methodologies for key experiments cited in probabilistic model development and validation for 505(b)(2) applications.
Protocol: Comparative Bioavailability PK Bridging Study with Probabilistic Analysis
1. Objective: To establish a scientific bridge between the Sponsor's 505(b)(2) product and the Reference Listed Drug (RLD) through a comparative pharmacokinetic (PK) study, and to analyze the results using a probabilistic model to quantify the likelihood of meeting regulatory criteria.
2. Experimental Design:
- Design: Single-dose, randomized, crossover study under fasting conditions.
- Subjects: N=36 healthy adult subjects (providing ~90% power to demonstrate bioequivalence for AUC with a 5% alpha error).
- Treatments: Test (T) 505(b)(2) product vs. Reference (R) RLD.
- Washout Period: At least 5 half-lives of the drug.
3. Procedures:
- Subjects are randomized to receive sequence TR or RT.
- Blood samples are collected pre-dose and at specified intervals up to 48 hours post-dose.
- Plasma concentrations are determined using a validated bioanalytical method (e.g., LC-MS/MS).
4. Data Analysis:
- Primary Endpoints: AUC~0-t~, AUC~0-∞~, and C~max~.
- Standard Statistical Analysis: Calculate geometric mean ratios (GMR) and 90% confidence intervals (CIs) for T/R. Standard bioequivalence is concluded if the 90% CI falls within 80.00%-125.00%.
- Probabilistic Analysis:
- A Bayesian hierarchical model is constructed using prior distributions informed by pre-clinical data and similar compounds.
- The model incorporates the observed PK data to generate a posterior distribution for the true GMR.
- The output is the Posterior Probability that the true GMR lies within the bioequivalence range (P~BE~). A P~BE~ > 0.90 is considered a high probability of success.
5. Model Verification:
- The Bayesian model is formally specified.
- Invariants are defined (e.g., "probability outputs must be between 0 and 1").
- Proof obligations are generated and discharged using the Rodin tool to ensure the model's logical and mathematical correctness [7].
Protocol: In Silico Simulation of Food Effect Using Physiologically Based Pharmacokinetic (PBPK) Modeling
1. Objective: To probabilistically predict the effect of food on the bioavailability of the 505(b)(2) product and inform the need for a clinical food-effect study.
2. Model Development:
- A PBPK model for the drug is developed and verified against existing RLD clinical data (fasting and fed states).
- The model incorporates system-dependent (human physiology) and drug-dependent (API permeability, solubility, dissolution) parameters.
3. Probabilistic Simulation:
- Key uncertain parameters (e.g., solubility in fed state) are defined as probability distributions rather than fixed values.
- A Monte Carlo simulation is run (e.g., n=1000 virtual trials) to predict the distribution of C~max~ and AUC GMRs (Fed/Fasted) for the 505(b)(2) formulation.
4. Output and Decision Rule:
- The simulation yields a probability distribution for the food effect.
- The probability (P~No-Effect~) that the GMRs remain within 80-125% is calculated.
- Decision: If P~No-Effect~ > 0.95, a clinical food-effect study may be waived. If P~No-Effect~ < 0.80, a clinical study is recommended. Intermediate probabilities trigger further formulation refinement.
The logical flow of this probabilistic, model-informed approach is depicted below.
The Scientist's Toolkit: Research Reagent Solutions
Successful implementation of a probabilistic 505(b)(2) program relies on a suite of essential materials and software tools.
Table 2: Essential Research Reagents and Tools for Probabilistic 505(b)(2) Development
| Item / Tool Name | Function / Application |
|---|---|
| Formulation Matrices | Pre-defined libraries of excipients and their compatibility data for developing new dosage forms (e.g., extended-release matrices, abuse-deterrent polymers). |
| In Vitro Dissolution Apparatus (USP I, II, IV) | To assess drug release profiles and establish an in vitro-in vivo correlation (IVIVC), a critical component for justifying biowaivers. |
| Validated Bioanalytical Method (e.g., LC-MS/MS) | For the precise and accurate quantification of drug and metabolite concentrations in biological samples from PK bridging studies. |
| Model-Informed Drug Development (MIDD) Platforms | Software for PBPK modeling, population PK/PD analysis, and exposure-response modeling to support scientific bridging and study waivers [53]. |
| Formal Verification Software (e.g., Rodin Platform, Event-B) | Tools for the formal modeling and verification of probabilistic algorithms, ensuring their mathematical correctness and reliability [7]. |
| Clinical Data Visualization & Analytics (e.g., JMP Clinical) | Interactive software for exploring clinical trial data, detecting trends and outliers, and performing safety and efficacy analyses to inform model inputs [55]. |
| Statistical Computing Environment (e.g., R, SAS, elluminate SCE) | Validated environments for conducting statistical analyses, including complex Bayesian modeling and simulation, with traceability for regulatory submissions [56]. |
Data Presentation and Analysis
Quantitative data from experiments and models must be synthesized for clear decision-making. The following table summarizes potential outputs from a probabilistic analysis of a critical quality attribute.
Table 3: Probabilistic Analysis of Simulated C~max~ Geo. Mean Ratio (GMR) for a 505(b)(2) Formulation
| Percentile | Simulated GMR (T/R) | Interpretation |
|---|---|---|
| 2.5th | 88.5% | Lower bound of the 95% prediction interval. |
| 50th (Median) | 98.2% | Most likely outcome. |
| 97.5th | 108.1% | Upper bound of the 95% prediction interval. |
| Probability that GMR is within 90.00%-111.11% | 96.7% | High confidence in achieving tighter acceptance criteria. |
| Probability that GMR is within 80.00%-125.00% | >99.9% | Virtual certainty of standard bioequivalence. |
This data presentation format allows researchers and regulators to assess not just a point estimate, but the full range of likely outcomes and the associated confidence, which is a cornerstone of a probabilistic approach to verification and validation.
Navigating Challenges: Common Pitfalls and Strategic Optimization
In the realm of probabilistic model verification and validation (V&V), practitioners navigate a critical tension between two fundamental pitfalls: over-simplification that omits crucial real-world phenomena, and unjustified complexity that introduces unnecessary computational burden and opacity. A probabilistic approach to V&V provides a mathematical framework to quantify and manage this trade-off, enabling researchers to make informed decisions about model structure and complexity while rigorously characterizing predictive uncertainty.
This balance is particularly crucial in safety-critical domains like drug development, where models must be both tractable for formal verification and sufficiently rich to capture essential biological dynamics. The following application notes provide structured methodologies and protocols for implementing probabilistic V&V frameworks that explicitly address this tension, supporting both regulatory compliance and scientific innovation in pharmaceutical research and development.
Quantitative Framework for Model Limitation Assessment
Characterization of Model Limitation Types
Table 1: Quantitative Profiles of Model Limitation Types
| Limitation Type | Key Indicators | Verification Challenges | Validation Challenges | Probabilistic Quantification Methods |
|---|---|---|---|---|
| Over-Simplification | - Overconfident predictions- Systematic residuals in calibration- Poor extrapolation performance | - False guarantees due to omitted variables- Overly broad assumptions | - Consistent underperformance on specific data subsets- Failure in edge cases | - Bayesian model evidence- Posterior predictive checks- Mismatch in uncertainty quantification |
| Unjustified Complexity | - High variance in predictions- Sensitivity to noise- Poor identifiability | - State space explosion- Intractable formal verification- Excessive computational demands | - Overfitting to training data- Poor generalization to new data | - Bayes factors- Watanabe-Akaike Information Criterion (WAIC)- Cross-validation metrics |
Probabilistic Metrics for Limitation Assessment
Table 2: Quantitative Metrics for Model Limitation Assessment
| Metric Category | Specific Metrics | Target Range | Interpretation for Limitation Assessment |
|---|---|---|---|
| Model Fit Metrics | - Bayesian R²- Watanabe-Akaike Information Criterion (WAIC)- Log pointwise predictive density | - R²: 0.7-0.9- Lower WAIC preferred- Higher density preferred | - Values outside range indicate poor fidelity- Significant differences between training/validation suggest over-complexity |
| Uncertainty Quantification | - Posterior predictive intervals- Calibration curves- Sharpness | - 95% interval should contain ~95% of observations- Calibration curve close to diagonal- Balanced sharpness | - Overly narrow intervals indicate overconfidence from simplification- Overly wide intervals suggest unjustified complexity |
| Model Comparison | - Bayes factors- Posterior model probabilities- Cross-validation scores | - Bayes factor >10 for strong evidence- Probability >0.95 for preferred model- Stable CV scores | - Quantifies evidence for simpler vs. more complex models- Guides model selection |
Experimental Protocols for Probabilistic V&V
Application Context: This protocol is adapted from unified probabilistic verification methodologies for vision-based autonomous systems [17] and is particularly relevant for complex pharmacological models with perceptual uncertainties or multiple interacting components.
Research Reagent Solutions:
- Probabilistic Model Checker: Software tools (e.g., PRISM, Storm) for formal verification of stochastic models [17]
- Interval Markov Decision Process (IMDP) Framework: Mathematical structure for representing uncertainty in transition probabilities [17]
- Bayesian Validation Tools: Statistical methods for quantifying model validity in new environments [17]
Methodology:
- System Abstraction:
- Map the continuous model to a discrete-state IMDP abstraction ℳ that overapproximates the concrete system with confidence α [17]
- Represent neural perception uncertainties or biological variabilities with confidence intervals on transition probabilities
- Ensure the abstraction contains the behavior distribution of the true system with probability 1-α
Verification Step:
- Formalize system-level temporal properties φ (e.g., "probability of adverse event never exceeds threshold")
- Employ probabilistic model checker to compute upper bound β on the chance that a trajectory falsifies φ [17]
- Generate frequentist-style guarantee: "With confidence α in the dataset, the underlying system produces safe trajectories with probability ≥1-β" [17]
Validation in Deployment Environment:
- Collect new data from deployment environment E'
- Construct belief on parameters of ME' based on new data using Bayesian methods
- Compute posterior probability 1-γ that new environment E' falls within uncertainty of ℳ [17]
- Establish nested guarantee: "With confidence 1-γ, system ME' satisfies property φ with probability 1-β" [17]
Protocol 2: Model Calibration and Uncertainty Guidance for Biogeochemical Modeling
Application Context: Adapted from Verra's VMD0053 guidance for agricultural land management [57], this protocol provides a robust framework for pharmacological model calibration, validation, and uncertainty assessment, particularly useful for complex biological systems with limited observability.
Research Reagent Solutions:
- Independent Modeling Expert (IME): Qualified external validator for objective model assessment [57]
- Process-Based Biogeochemical Model: Mechanistic models representing biological processes [57]
- Uncertainty Quantification Tools: Statistical methods for propagating parameter and structural uncertainties [57]
Methodology:
- Model Calibration:
- Establish calibration dataset representing the spectrum of expected operating conditions
- Define objective function(s) quantifying mismatch between predictions and observations
- Implement multi-step calibration procedure:
- Preliminary sensitivity analysis to identify influential parameters
- Formal parameter estimation using Bayesian or likelihood-based methods
- Validation against withheld portion of calibration dataset
Model Validation:
- Collect independent validation dataset not used in calibration
- Perform quantitative comparison between model predictions and validation observations
- Assess both point predictions and uncertainty estimates using statistical measures
- Document performance metrics and any systematic biases
Uncertainty Characterization:
- Quantify parameter uncertainty via posterior distributions (Bayesian) or confidence intervals (frequentist)
- Assess model structural uncertainty through multi-model ensemble approaches
- Evaluate scenario uncertainty for future projections or extrapolations
- Propagate uncertainties through to model outputs using appropriate methods
Protocol 3: Probabilistic Audits for Verifiable Training and Outcome Improvement
Application Context: This protocol adapts recent advances in verifiable decentralized learning [58] to pharmacological model development, providing mechanisms to ensure both correct training procedures and genuine model improvement while managing computational costs.
Research Reagent Solutions:
- Proof-of-Improvement (PoI) Framework: Lightweight evaluation audit that statistically certifies milestone-based gains [58]
- Economic Security Mechanisms: Cryptographic commitments and incentive structures for audit integrity [58]
- Statistical Power Analysis Tools: Methods for determining appropriate audit sample sizes [58]
Methodology:
- Process Verification via Probabilistic Audits:
- Provers commit to each training step with cryptographic commitments
- Verifier committees audit a small, random fraction of steps (reducing verification compute by >95% compared to full replication) [58]
- Derive tight detection-cost frontier to minimize verification overhead while maintaining security
Outcome Verification via Proof-of-Improvement:
- Define clinically meaningful improvement metrics (e.g., perplexity reduction, accuracy gain)
- Statistically certify milestone-based gains on a committed dataset [58]
- Implement with high statistical power at minimal computational cost
Integrated Verification Reporting:
- Combine process and outcome verification results
- Generate comprehensive certificates of model integrity and improvement
- Document audit methodology, sample sizes, and statistical power
Integrated Workflow for Model V&V
The following diagram integrates the three protocols into a comprehensive workflow for managing model complexity throughout the development lifecycle:
The tension between over-simplification and unjustified complexity represents a fundamental challenge in pharmacological model development. The probabilistic V&V frameworks presented herein provide structured methodologies for navigating this trade-off, enabling researchers to build models that are both sufficiently rich to capture essential biological phenomena and sufficiently tractable for rigorous verification and validation. By implementing these protocols, drug development professionals can enhance model credibility, support regulatory submissions, and ultimately accelerate the delivery of safe and effective therapeutics.
Overcoming Organizational and Resource Barriers to MIDD Adoption
Model-Informed Drug Development (MIDD) represents a paradigm shift in pharmaceuticals, leveraging quantitative models to streamline development and inform decision-making. A probabilistic approach to model verification and validation (V&V) is fundamental to this framework, ensuring that models are not only technically correct but also robust and reliable for predicting real-world outcomes. This approach moves beyond deterministic checks, incorporating uncertainty quantification and rigorous assessment of model performance under varying conditions. However, the adoption of these sophisticated methodologies is often hampered by significant organizational and resource barriers. This document outlines these challenges and provides detailed application notes and protocols to facilitate their successful integration into drug development pipelines.
Systematic Analysis of Adoption Barriers
The successful implementation of any complex technological framework, including MIDD, is influenced by a constellation of factors. A systematic review of digital health tool adoption among healthcare professionals identified and categorized 125 barriers and 108 facilitators, which can be directly mapped to the MIDD context [59]. These were consolidated into five key domains: Technical, User-related, Economical, Organizational, and Patient-related. The following table synthesizes these findings with known challenges in MIDD and probabilistic V&V.
Table 1: Categorized Barriers to MIDD and Probabilistic V&V Adoption
| Category | Specific Barriers | Relevance to MIDD & Probabilistic V&V |
|---|---|---|
| Technical & Resource | Need for additional training; Time consumption; Poor interoperability of systems and data [59] [60]. | Steep learning curve for probabilistic programming (e.g., Stan, PyMC3); Computational demands of Markov Chain Monte Carlo (MCMC) methods; Lack of standardized data formats for model ingestion. |
| User-related & Cultural | Resistance to change; Lack of buy-in; Concerns over impact on autonomy and workflow [59] [61] [62]. | Cultural preference for traditional, deterministic approaches; "Black box" mistrust of complex models; Perceived threat to expert judgment from data-driven recommendations. |
| Organizational & Leadership | Lack of a clear and cohesive vision from senior leadership; Poor governance; Weak sponsorship and communication gaps [59] [61] [62]. | Absence of a cross-functional MIDD strategy; Inadequate decision-making frameworks for model-informed choices; Lack of visible executive champions for quantitative approaches. |
| Economic & Infrastructural | High initial costs; Financial constraints; Unreliable infrastructural support [59] [63]. | Significant investment required for high-performance computing (HPC) resources; Costs associated with hiring specialized talent; Unstable or slow computational networks hindering complex simulations. |
A critical, often-overlooked challenge is change fatigue, identified as a high-impact barrier where too many concurrent initiatives exceed an organization's capacity to absorb change [62]. Rolling out a probabilistic V&V framework amidst other major changes can lead to disengagement and failure.
Application Notes and Experimental Protocols for Probabilistic V&V
To overcome these barriers, a structured, protocol-driven approach is essential. The following section provides a detailed methodology for key activities.
Protocol 1: Establishing a Probabilistic Model V&V Framework
Objective: To create a standardized, organization-wide protocol for the verification and validation of MIDD models using a probabilistic paradigm.
Background: Probabilistic V&V assesses not just if a model is "correct," but quantifies the confidence in its predictions under uncertainty. Verification ensures the model is implemented correctly, while validation assesses its accuracy against empirical data.
Table 2: Research Reagent Solutions for Probabilistic V&V
| Reagent / Tool | Function / Explanation |
|---|---|
| Probabilistic Programming Language (e.g., Stan, PyMC3) | Enables specification of complex Bayesian statistical models and performs efficient posterior inference using algorithms like MCMC and Variational Inference. |
| High-Performance Computing (HPC) Cluster | Provides the necessary computational power to run thousands of complex, stochastic simulations and MCMC sampling in a feasible timeframe. |
| Modeling & Simulation Software (e.g., R, Python with NumPy/SciPy) | The core environment for building and testing models, performing data analysis, and visualizing results. |
| Standardized Datasets (e.g., PK/PD, Clinical Trial Data) | Curated, high-quality datasets are the "reagents" against which models are validated. They must be representative of the target population. |
| Containerization Technology (e.g., Docker, Singularity) | Ensures computational reproducibility by packaging the model code, dependencies, and environment into a single, portable unit. |
Methodology:
- Verification Phase:
- Code Review: Implement peer-review of model code, focusing on the correct implementation of prior distributions, likelihood functions, and sampling algorithms.
- Unit Testing: Develop tests for individual model components (e.g., PK ODE solver) to ensure they function as intended in isolation.
- Predictive Checks:
- Prior Predictive Checks: Simulate data using only the prior distributions to assess if they are realistic and scientifically plausible.
- Posterior Predictive Checks: Simulate new data from the fitted model and compare it to the observed data. A well-specified model will generate data that looks similar to the actual observations.
- Validation Phase:
- Internal Validation: Use techniques like k-fold cross-validation on the available dataset to assess model robustness and check for overfitting.
- External Validation: Test the model's predictive performance on a completely new, independent dataset not used during model development. This is the gold standard.
- Face Validation: Engage domain experts (e.g., clinical pharmacologists, physicians) to review model assumptions, structure, and outputs for biological and clinical plausibility.
The workflow below illustrates the logical sequence and iterative nature of this probabilistic V&V process.
Protocol 2: A Change Management Plan for MIDD Implementation
Objective: To provide a detailed protocol for leading the organizational change required to overcome cultural and leadership barriers to MIDD adoption.
Background: Technical excellence alone is insufficient. A 2025 report highlighted that 44% of respondents rated change fatigue as a high-impact barrier, making it a critical risk to manage [62]. Successful adoption requires a disciplined change management process.
Methodology:
- Craft a Change Brief: Create a concise document stating the "why," target outcomes, scope, key owners, and 3-5 measurable success metrics for MIDD adoption. This removes ambiguity and serves as a single source of truth [62].
- Activate Visible Sponsorship: Secure a C-level sponsor who is responsible for championing the initiative. Their role includes leading town halls, unblocking issues, and consistently communicating the vision. Tie their KPIs to adoption metrics, not just project delivery [61] [62].
- Co-create with Impacted Roles: Involve key researchers, clinicians, and regulatory strategists early through pilot projects. Gather their feedback on workflows and show visible fixes that result from their input. This builds credibility and creates peer advocates [62].
- Deliver Contextual Training: Move beyond one-time training events. Provide role-based onboarding and embed performance support (e.g., in-app guidance, sandbox environments) directly into the workflow to shorten time to proficiency [59] [62].
- Measure and Course-Correct: Track a small set of leading indicators (e.g., tool usage, training completion) and lagging indicators (e.g., model-influenced decisions, cycle time reduction). Review these KPIs regularly and adjust the strategy accordingly [62].
Overcoming the organizational and resource barriers to MIDD adoption, particularly within the rigorous framework of probabilistic V&V, requires a dual-focused strategy. It demands both technical excellence, achieved through robust and standardized experimental protocols, and organizational agility, fostered by a deliberate and empathetic change management plan. By systematically addressing the technical, cultural, and leadership challenges outlined herein, research organizations can build the capability and credibility needed to fully leverage the power of model-informed drug development, ultimately leading to more efficient processes and safer, more effective therapeutics for patients.
Model-informed drug development (MIDD) leverages quantitative approaches to facilitate drug development and regulatory decision-making. A probabilistic approach to model verification and validation (V&V) is becoming critical, moving beyond deterministic "pass/fail" checks to a framework that quantifies confidence, uncertainty, and risk. This paradigm shift allows for a more nuanced assessment of a model's predictive power under conditions of uncertainty and distribution shift, which is essential for regulatory acceptance. This document provides detailed application notes and protocols for preparing probabilistic V&V evidence for interactions with the U.S. Food and Drug Administration (FDA) and the European Medicines Agency (EMA).
Regulators are increasingly advocating for advanced methodologies. The FDA's New Alternative Methods Program aims to spur the adoption of methods that can replace, reduce, and refine animal testing, highlighting the role of computational models. A key aspect is the qualification process, where a tool is evaluated for a specific context of use (COU), defining the boundaries within which available data justify its application [64]. Similarly, the EMA has emphasized the need for optimal tools to assess groundbreaking therapies through its Regulatory Science Strategy to 2025, which focuses on catalyzing the integration of science and technology in medicine development [65]. A unified V&V methodology aims to provide flexible, end-to-end guarantees that adapt to out-of-distribution test-time conditions, offering a rigorous yet practical safety assurance for complex biological models [17].
Regulatory Landscape and Quantitative Submissions Analysis
Understanding recent regulatory trends is paramount for successful agency interactions. Regulatory policies are dynamic, and a probabilistic approach must be informed by the current landscape of submissions and precedents.
Recent FDA Policy Shifts
The FDA under Commissioner Dr. Martin Makary has initiated several new policies that impact regulatory strategy. Key developments include:
- Enhanced Transparency: The FDA has begun publishing over 200 complete response letters (CRLs) for drug and biological products from 2020-2024. This unprecedented transparency offers insights into the agency's decision-making rationale and common deficiencies, which sponsors can use to inform their own applications [66].
- National Priority Voucher (CNPV) Program: This pilot program may fast-track reviews for drugs that advance national health priorities. Notably, the Commissioner has suggested that "equalizing" drug costs between the U.S. and other countries could be a favorable factor, a significant step into pricing considerations for an agency traditionally focused on safety and efficacy [66].
- Focus on Clinical Trial Transparency: FDA's Center for Drug Evaluation and Research (CDER) is emphasizing compliance with clinical trial registration and results reporting on ClinicalTrials.gov, viewing it as an ethical obligation [67].
Quantitative Analysis of PBPK Submissions at FDA CBER
A landscape review of PBPK modeling submissions to the FDA's Center for Biologics Evaluation and Research (CBER) provides a quantitative benchmark for regulatory preparedness. The following table summarizes submissions and interactions from 2018 to 2024 [68].
Table 1: CBER Experience with PBPK Modeling & Simulation (2018-2024)
| Category | Specific Type | Number of Submissions/Interactions |
|---|---|---|
| Overall Trend | Increasing number over time | 26 total |
| Submission Type | Investigational New Drug (IND) | 10 |
| Pre-IND Meetings | 8 | |
| Biologics License Application (BLA) | 1 | |
| Other (INTERACT, MIDD, Type V DMF) | 7 | |
| Product Category | Gene Therapy Products | 8 |
| Plasma-Derived Products | 3 | |
| Vaccines | 1 | |
| Cell Therapy Product | 1 | |
| Other (small molecules, bacterial lysates) | 5 | |
| Therapeutic Focus | Rare Diseases | 11 (of 18 products) |
This data demonstrates that PBPK and other mechanistic models are actively used across biological products, with a strong presence in rare diseases and early-stage development (pre-IND and IND). This establishes a precedent for submitting probabilistic models to regulators.
Experimental Protocols for Model Verification & Validation
A robust probabilistic V&V framework requires structured methodologies. The protocols below are designed to generate evidence that meets regulatory standards for credibility.
Protocol 1: Risk-Based Credibility Assessment
Objective: To establish the credibility of a computational model for its specific Context of Use (COU) as per FDA and EMA expectations [64] [69]. Materials: Model code, input data, validation dataset, high-performance computing (HPC) resources. Procedure:
- Define Context of Use: Formally specify the question the model will answer, its boundaries, and the impact of its output on the decision. This is the foundation for all subsequent V&V.
- Assess Model Risk: Categorize model risk as low, medium, or high based on the COU. A higher-risk model (e.g., one predicting clinical efficacy) requires more extensive V&V.
- Verification (Is the model implemented correctly?):
- Code Verification: Use static analysis and unit testing to ensure the code is error-free.
- Numerical Verification: Perform grid convergence studies and check for conservation of mass/energy.
- Uncertainty Quantification: Propagate input uncertainties (e.g., via Monte Carlo sampling) to quantify their impact on outputs.
- Validation (Is the model an accurate representation?):
- Conduct Bayesian Validation: Update prior beliefs about model parameters with new data from the deployment environment to compute a posterior probability
(1-γ)that the real-world system falls within the model's uncertainty bounds [17]. - Calculate a frequentist safety guarantee: Using a probabilistic model checker, compute an upper bound
βon the probability of a safety requirement violation. - The combined result is a nested probabilistic guarantee: "With confidence
(1-γ), the system satisfies the safety property with probability at least(1-β)" [17].
- Conduct Bayesian Validation: Update prior beliefs about model parameters with new data from the deployment environment to compute a posterior probability
- Documentation: Compile a comprehensive report linking all evidence to the COU and risk assessment.
Protocol 2: PBPK Model Development and Qualification for Pediatric Extrapolation
Objective: To develop a PBPK model for a therapeutic protein (e.g., Fc-fusion protein) to support pediatric dose selection, as demonstrated in the ALTUVIIIO case [68]. Materials: Physiological parameters, in vitro ADME data, adult and pediatric clinical PK data (if available), PBPK software platform (e.g., GastroPlus, Simcyp). Procedure:
- Model Building: Develop a minimal PBPK model structure incorporating key clearance mechanisms (e.g., FcRn recycling). Populate with system-dependent (physiological) and drug-dependent (in vitro) parameters.
- Model Qualification with a Surrogate: Use clinical PK data from a reference product (e.g., ELOCTATE) with a similar mechanism to validate the model's predictive performance in both adults and children. Optimize age-dependent parameters (e.g., FcRn abundance) using the reference data.
- Acceptance Criteria: Define prediction error boundaries (e.g., ±25% for Cmax and AUC) as a probabilistic validation metric [68].
- Probabilistic Simulation: Use the qualified model to simulate the exposure of the new investigational product (e.g., ALTUVIIIO) in a virtual pediatric population. Predict the proportion of the population maintaining target exposure (e.g., FVIII activity >20 IU/dL) over a dosing interval.
- Regulatory Submission Package: Include the model structure, input parameters, qualification results against the reference product, and simulation outcomes with uncertainty intervals to justify the proposed pediatric dose.
Visualizing the Probabilistic V&V Workflow
The following diagram illustrates the integrated verification, validation, and regulatory interaction pathway, synthesizing the concepts from the experimental protocols.
Probabilistic V&V and Regulatory Pathway
The Scientist's Toolkit: Essential Research Reagent Solutions
Successful implementation of the protocols requires specific tools and reagents. The following table details key materials and their functions in the context of probabilistic V&V for regulatory submissions.
Table 2: Essential Research Reagent Solutions for Probabilistic Model V&V
| Tool/Reagent | Function in Probabilistic V&V | Regulatory Context |
|---|---|---|
| PBPK Software Platform (e.g., GastroPlus, Simcyp) | Provides a mechanistic framework to simulate ADME processes; enables virtual population trials and sensitivity analysis. | Accepted in INDs/BLAs for DDI, pediatric, and formulation risk assessment [68]. |
| Interval Markov Decision Process (IMDP) Tools | Creates abstractions that over-approximate system behavior with confidence α, accounting for perceptual and dynamic uncertainty [17]. |
Emerging tool for verifying AI/ML-based autonomous systems; foundational for rigorous safety guarantees. |
| Probabilistic Model Checker (e.g., PRISM, Storm) | Automatically verifies temporal logic properties on probabilistic models (e.g., IMDPs) to compute safety probability bounds β [17]. |
Used in research to generate quantifiable, worst-case safety certificates for closed-loop systems. |
| Virtual Population (ViP) Models | A set of high-resolution anatomical models used as a gold standard for in silico biophysical modeling [64]. | Cited in over 600 CDRH premarket applications; demonstrates regulatory acceptance of virtual evidence. |
| Microphysiological Systems (Organ-on-Chip) | 3D in vitro models that emulate human organ physiology for safety and efficacy testing; a key New Alternative Method [64]. | Subject of the first ISTAND program submission; potential to reduce animal testing. |
| Bayesian Inference Tools (e.g., Stan, PyMC) | Updates model parameter beliefs with new data to compute posterior validity probability (1-γ) for new environments [17]. |
Core to the Bayesian validation protocol, bridging the gap between lab model and real-world deployment. |
Navigating FDA and EMA interactions for complex, model-based submissions requires a strategic shift towards probabilistic verification and validation. By adopting the structured application notes and protocols outlined herein—grounding models in a defined COU, implementing rigorous risk-based credibility assessments, leveraging quantitative submission data for planning, and generating nested probabilistic guarantees—sponsors can build robust evidence packages. This approach not only aligns with current regulatory initiatives but also proactively addresses the challenges of validating intelligent systems in the face of uncertainty, thereby optimizing development and accelerating patient access to safe and effective therapies.
In critical research fields such as drug development, the challenge of deriving robust, verifiable conclusions from limited datasets is a significant constraint. A probabilistic approach to model verification and validation provides a rigorous framework for navigating this challenge, transforming data scarcity from a liability into a parameter of explicit uncertainty. This paradigm shift is essential for maintaining scientific integrity when comprehensive data collection is infeasible, as is often the case in rare diseases, early-stage clinical trials, and complex biological systems. By quantifying uncertainty rather than ignoring it, researchers can make informed decisions, prioritize resource allocation, and build models that accurately represent the known boundaries of knowledge. These protocols outline practical strategies for ensuring data quality, applying statistical techniques suited for small samples, and implementing probabilistic verification within the drug development workflow, providing a structured methodology for researchers and scientists.
Foundational Data Quality Framework
High-quality data is the non-negotiable foundation for any analysis, especially when dataset size is limited. Poor quality data can result in additional spend of $15M in average annual costs for organizations and affects all levels from operational processing to strategic decision-making [70]. A structured data quality framework is therefore essential.
The Six Core Dimensions of Data Quality
Data quality is a multi-dimensional construct. Measuring these dimensions allows for targeted improvement and establishes fitness for use in probabilistic modeling [70].
Table: The Six Core Dimensions of Data Quality for Research Datasets
| Dimension | Definition | Assessment Method | Impact on Probabilistic Models |
|---|---|---|---|
| Accuracy | The degree to which data correctly represents the real-world object or event it is intended to model [70]. | Verification against an authoritative source or the actual entity [70]. | Directly biases model parameters, leading to inaccurate predictions and invalid uncertainty quantification. |
| Completeness | The extent to which data is sufficient to deliver meaningful inferences and decisions without missing attributes [70]. | Checking for mandatory fields, null values, and missing values [70]. | Missing data increases uncertainty and can lead to overfitting if not handled explicitly within the model. |
| Consistency | The degree to which data stored and used across multiple instances or systems is logically aligned [70]. | Planned testing across multiple datasets or records for logical conflicts [70]. | Inconsistencies violate the assumption of a single data-generating process, corrupting the probabilistic model's likelihood function. |
| Validity | The degree to which data conforms to the specified syntax (format, type, range) of its definition [70]. | Application of business rules or formal schema validation (e.g., regular expressions for ZIP codes) [70]. | Invalid data points are often outliers that can skew the analysis and distort the estimated posterior distribution. |
| Uniqueness | The extent to which data is recorded only once within a dataset to avoid unnecessary duplication [70]. | Identification of duplicates or overlaps through data profiling and deduplication processes [70]. | Duplicate records artificially inflate the apparent sample size, leading to underestimated confidence intervals and overconfident predictions. |
| Timeliness | The degree to which data is available for use when required and is up-to-date for the task at hand [70]. | Checking the recency of the data and the time elapsed since last update [70]. | Stale data may not reflect the current system being modeled, rendering the probabilistic model's outputs irrelevant. |
Protocol for Data Quality Assurance and Cleaning
This protocol provides a step-by-step methodology for preparing a limited dataset for analysis, minimizing the introduction of errors from the data itself [3].
Protocol 2.2: Quantitative Data Quality Assurance
Objective: To systematically clean and quality-assure a research dataset to ensure it meets the standards required for robust probabilistic analysis.
Materials:
- Raw research dataset
- Statistical software (e.g., R, Python with pandas)
- Data documentation and codebooks
Procedure:
Check for Duplications:
- Identify and remove identical copies of data, leaving only unique participant records.
- Rationale: Prevents artificial inflation of the sample size, which is critical for accurate uncertainty estimation in small datasets [3].
Assess and Handle Missing Data:
- Distinguish between data that is missing (a response was expected but omitted) and not relevant (e.g., "not applicable") [3].
- Calculate the percentage of missing data per variable and per participant.
- Use a statistical test like Little's Missing Completely at Random (MCAR) test to determine the pattern of missingness [3].
- Decision Point:
- If data is MCAR and the percentage of missingness for a participant exceeds a pre-defined threshold (e.g., 50%), consider removing that participant.
- If data is not MCAR, employ advanced statistical techniques such as Multiple Imputation or Maximum Likelihood estimation to handle the missing data, as simple deletion can introduce bias [3].
Identify Anomalies (Outliers):
- Run descriptive statistics (minimum, maximum, mean) for all measures.
- Visually examine distributions using box plots and histograms to detect values that deviate from expected patterns (e.g., a Likert score outside its possible range) [3] [71].
- Investigate the source of anomalies to determine if they are errors (to be corrected or removed) or genuine, but extreme, values (to be retained) [3].
Validate Data Integrity and Summation:
Establish Psychometric Properties (if applicable):
- For datasets involving standardized instruments, report psychometric properties like reliability.
- Calculate Cronbach's alpha to assess internal consistency; scores > 0.7 are generally considered acceptable [3].
- If calculation for the study sample is not possible, report established psychometric properties from similar studies.
Diagram: Data Quality Assurance Workflow. This flowchart outlines the sequential protocol for cleaning a research dataset, from initial raw data to an analysis-ready state.
Statistical Techniques for Limited Data
When the quantity of data is fixed and limited, specialized statistical techniques are required to maximize the information extracted and properly quantify uncertainty.
Exploratory Data Analysis (EDA) for Characterizing Limited Data
EDA is the critical first step after data cleaning, involving the visual and statistical summarization of the dataset's main characteristics. It is essential for understanding the structure of a small dataset, identifying patterns, and uncovering potential outliers or anomalies that may have a disproportionate influence [71].
Protocol 3.1: Exploratory Data Analysis for a Limited Dataset
Objective: To gain an in-depth understanding of a limited dataset's distribution, variable relationships, and potential issues before formal modeling.
Materials:
- Cleaned dataset from Protocol 2.2
- Statistical software with visualization capabilities (e.g., Python with Seaborn/Matplotlib, R with ggplot2)
Procedure:
Visualize Variable Distributions:
- Create histograms or kernel density estimates (KDE) for continuous variables (e.g., age, biomarker concentration). This reveals the shape of the distribution (normal, skewed), central tendency, and spread [71].
- Create bar charts for categorical variables (e.g., treatment group, genotype).
Assess Inter-Variable Relationships:
- For two continuous variables, generate a scatter plot. For example, plot "Hours Studied" against "Exam Score" to visually assess correlation and form of relationship [71].
- For a continuous variable across categories, use box plots or violin plots. These visualize the median, quartiles, and range, and help identify potential outliers within groups [71].
Test for Normality of Distribution:
- This step is crucial for selecting appropriate statistical tests later.
- Calculate skewness and kurtosis. Values of ±2 for both are generally considered acceptable evidence of normality [3].
- Perform formal statistical tests like the Shapiro-Wilk test (preferred for smaller samples) or the Kolmogorov-Smirnov test [3].
- Decision Point: If data significantly deviates from normality, non-parametric statistical tests should be prioritized for inference.
Bootstrapping for Estimating Uncertainty
Bootstrapping is a powerful resampling technique that allows researchers to estimate the sampling distribution of a statistic (e.g., mean, median, regression coefficient) by repeatedly sampling with replacement from the observed data. It is exceptionally valuable for small datasets and complex statistics where theoretical formulas for confidence intervals may not be available or reliable [71].
Protocol 3.2: Bootstrapping for Confidence Intervals
Objective: To estimate the uncertainty (via a confidence interval) of a population parameter using only the data from a limited sample.
Materials:
- Cleaned dataset
- Statistical software (e.g., Python with NumPy/SciPy, R with boot package)
Procedure:
Define the Parameter of Interest:
- Identify the statistic you wish to estimate (e.g., the mean time to complete a task, the AUC of a pharmacokinetic profile).
Generate Bootstrap Samples:
- Set the number of bootstrap iterations,
B(typicallyB = 1000or more). - For each iteration
iin1...B:- Create a bootstrap sample by randomly selecting
Nobservations from the original dataset with replacement, whereNis the size of the original dataset [71]. - Calculate the statistic of interest for this bootstrap sample, denoted
θ_i.
- Create a bootstrap sample by randomly selecting
- Set the number of bootstrap iterations,
Analyze the Bootstrap Distribution:
- The collection of
Bstatistics (θ_1, θ_2, ..., θ_B) forms the bootstrap distribution. - This distribution approximates the sampling distribution of the statistic.
- The collection of
Calculate the Confidence Interval:
- Use the percentile method to construct a 95% confidence interval:
- Find the 2.5th percentile and the 97.5th percentile of the bootstrap distribution.
- The interval between these percentiles is the 95% bootstrap confidence interval for the parameter [71].
- Use the percentile method to construct a 95% confidence interval:
Diagram: Bootstrapping Process. This diagram illustrates the iterative process of bootstrapping, from resampling the original data to forming a distribution of the statistic and deriving a confidence interval.
Probabilistic Frameworks for Decision-Making in Drug Development
The probabilistic paradigm formalizes the use of existing knowledge and explicitly accounts for uncertainty, making it exceptionally powerful for decision-making with limited new data.
Bayesian Methods for Incorporating Prior Evidence
Bayesian statistics answer direct questions about the probability of a hypothesis given the data, P(H | D), by combining prior knowledge (D_0) with new experimental data (D_N). This is in contrast to frequentist methods, which calculate the probability of observing the data given a hypothesis, P(D | H) [72]. This approach is naturally suited to settings where data accumulates over time, as in clinical development.
Protocol 4.1: Bayesian Analysis for a Clinical Trial Endpoint
Objective: To update the belief about the effectiveness of a new drug by combining prior information with data from a new, potentially small, clinical study.
Materials:
- Prior distribution (based on pre-clinical data, earlier phases, or literature)
- New clinical trial data
- Computational software for Bayesian analysis (e.g., R with RStan, PyMC3 in Python)
Procedure:
Define a Prior Distribution:
- Quantify existing knowledge or skepticism about the parameter of interest (e.g., the difference in response rates between drug and placebo). For example, a skeptical prior might be centered on zero with a narrow variance [72].
Construct the Likelihood Function:
- This represents the new data. For a binary endpoint, the likelihood is typically a Binomial distribution.
Compute the Posterior Distribution:
- Apply Bayes' Theorem:
Posterior ∝ Prior × Likelihood. - Use computational methods like Markov Chain Monte Carlo (MCMC) sampling to derive the full posterior distribution, which represents the updated knowledge about the parameter [72].
- Apply Bayes' Theorem:
Make Probabilistic Decisions:
- Calculate the probability that the drug effect is greater than a clinically relevant threshold (e.g.,
P(Response Rate > 20%)). - This probability can be directly used for Go/No-Go decisions in drug development pipelines [72].
- Calculate the probability that the drug effect is greater than a clinically relevant threshold (e.g.,
The Probability of Pharmacological Success (PoPS) Framework
The PoPS is an evidence-based, quantitative framework used in early drug development to estimate the probability that a molecule will achieve adequate pharmacology in most patients while minimizing safety risk, given all current uncertainties [38].
Protocol 4.2: Implementing a PoPS Analysis
Objective: To integrate multi-source data and uncertainties into a single probability to inform molecule progression decisions.
Materials:
- Pharmacokinetic (PK) and pharmacodynamic (PD) data (in vitro, in vivo)
- Exposure-response models for efficacy and safety
- Defined success criteria for pharmacology and safety endpoints
- Virtual population simulation software
Procedure:
Select Endpoints and Define Success Criteria:
- Efficacy Criterion: Define the level of a pharmacological endpoint (e.g., target inhibition, pathway modulation) expected to produce clinical efficacy, and the proportion of patients required to achieve it (e.g.,
K > 80% inhibition for >70% of subjects) [38]. - Safety Criterion: Define an acceptable exposure limit based on toxicology data (e.g., NOAEL from animal studies) and the proportion of patients allowed to exceed it [38].
- Efficacy Criterion: Define the level of a pharmacological endpoint (e.g., target inhibition, pathway modulation) expected to produce clinical efficacy, and the proportion of patients required to achieve it (e.g.,
Quantify Uncertainties:
- Model uncertainty in exposure-response parameters (e.g.,
CL ~ d1(λ1),Vd ~ d2(λ2)). - Model uncertainty in the success criteria themselves (e.g.,
K ~ d3(λ3)) [38].
- Model uncertainty in exposure-response parameters (e.g.,
Simulate Virtual Populations:
- For
Miterations (e.g., 500):- Sample a set of model parameters from their uncertainty distributions.
- Simulate a virtual population of
Nsubjects (e.g., 1000), generating PK/PD endpoints for each [38].
- For
Compute PoPS:
- For each virtual population, check if the pre-defined efficacy and safety criteria are jointly met.
- The PoPS is calculated as
PoPS = M' / M, whereM'is the number of virtual populations meeting all success criteria [38]. This single metric encapsulates the overall benefit-risk assessment.
Table: Essential Research Reagent Solutions for Probabilistic Analysis
| Reagent / Tool | Function / Purpose | Application Context |
|---|---|---|
| MCMC Sampler (e.g., Stan, PyMC) | Computationally approximates complex posterior distributions in Bayesian analysis. | Essential for Protocol 4.1 (Bayesian Analysis) when analytical solutions are intractable. |
| PK/PD Modeling Software (e.g., NONMEM, Monolix) | Develops mathematical models describing drug concentration (PK) and effect (PD) over time. | Core to building the exposure-response models required for Protocol 4.2 (PoPS Analysis). |
| Virtual Population Simulator | Generates in-silico patients with realistic physiological and demographic variability. | Used in PoPS analysis (Protocol 4.2) to predict outcomes in a target patient population. |
| Data Profiling Tool | Automates the assessment of data quality dimensions across a dataset. | Supports the initial stages of Protocol 2.2 (Data Quality Assurance) by identifying anomalies and missingness. |
Bootstrapping Library (e.g., boot in R, SciPy in Python) |
Provides functions to easily implement the resampling and calculation procedures in Protocol 3.2. | Key for uncertainty quantification with limited data without strong distributional assumptions. |
Integrated Protocol for Model Verification with Limited Data
This final protocol integrates the concepts of data quality, statistical techniques, and probabilistic thinking into a cohesive workflow for model verification.
Protocol 5: Integrated Probabilistic Verification of a Predictive Biomarker Model
Objective: To verify the predictive performance of a biomarker model for patient stratification, acknowledging the limitations of a small training dataset.
Materials:
- Limited dataset of biomarker measurements and patient outcomes.
- Computational environment for model training and probabilistic analysis.
Procedure:
Data Foundation & Quality Assurance:
- Execute Protocol 2.2 to clean and profile the biomarker and outcome data. Pay special attention to the accuracy (assay validation) and completeness of the biomarker measurements.
Exploratory Analysis & Bootstrapping:
- Execute Protocol 3.1 to visualize the relationship between the biomarker and the outcome.
- Execute Protocol 3.2 to estimate the confidence interval for the model's performance metric (e.g., the Area Under the ROC Curve, AUC). This provides a robust measure of performance uncertainty.
Probabilistic Model Verification:
- Frequentist Cross-Validation: Perform
k-foldcross-validation, reporting the mean and bootstrapped confidence interval of the performance metric. - Bayesian Verification: Treat the model's performance as an uncertain parameter. Use a prior distribution (e.g., based on biological plausibility) and update it with the cross-validated likelihood to compute a posterior distribution for the performance metric (applying principles from Protocol 4.1). This yields a direct probability statement about the model's adequacy (e.g.,
P(AUC > 0.7 | Data)).
- Frequentist Cross-Validation: Perform
Reporting and Decision Framework:
- Report the verified performance not as a single point estimate, but as a probability distribution.
- For a decision threshold (e.g.,
AUC > 0.65), calculate the probability this threshold is met. This integrated, probabilistic report provides a transparent and rigorous foundation for deciding whether to proceed with the biomarker.
The deployment of AI/ML models in drug development represents a paradigm shift in how researchers approach complex biological problems. However, a core challenge persists: the assumption that data distributions remain static between model training and real-world deployment is often violated, leading to degraded performance and potential safety risks. This phenomenon, known as distributional shift, necessitates robust probabilistic verification and validation (V&V) frameworks to ensure model reliability throughout the product lifecycle [73] [74].
Within the context of a broader thesis on probabilistic approaches to model V&V, this document establishes that distribution shifts are not merely operational inconveniences but fundamental challenges to the validity of safety assurances. A unified V&V methodology is therefore essential, producing safety guarantees that can adapt to out-of-distribution test-time conditions, thereby bridging the gap between theoretical verification and practical deployment [17].
Understanding Distributional Shifts: A Typology and Challenges
Distributional shifts occur when the statistical properties of input data or the relationship between inputs and outputs change between the training and deployment environments. In high-stakes fields like drug development, these shifts can compromise model integrity and patient safety [73]. The table below summarizes the three primary types of distributional shifts, their characteristics, and associated risks.
Table 1: A Typology of Distributional Shifts in AI/ML Models
| Shift Type | Formal Definition | Real-World Example | Primary Risk |
|---|---|---|---|
| Covariate Shift | ( P{train}(x) \neq P{test}(x) )( P{train}(y|x) = P{test}(y|x) ) | A manufacturing process is scaled up, leading to extended equipment usage times, higher operating temperatures, and lower lubricant levels—all altering the input feature distribution [73]. | Model fails to generalize despite unchanged input-output relationships, leading to inaccurate predictions on the new covariate distribution. |
| Label Shift | ( P{train}(y) \neq P{test}(y) )( P{train}(x|y) = P{test}(x|y) ) | A milling machine more prone to breakdowns is monitored, increasing the frequency of the "failure" label in the deployment data [73]. | Model's prior probability estimates become incorrect, skewing predictions and reducing accuracy for the now-more-prevalent class. |
| Concept Shift | ( P{train}(y|x) \neq P{test}(y|x) ) | A new maintenance routine is implemented, allowing machines to operate safely for longer periods, thus changing the fundamental relationship between "Usage Time" and "Machine Health" [73]. | The model's learned mapping from inputs to outputs becomes obsolete, rendering its decision logic invalid. |
A significant verification challenge is the fragility of vision-based observers and other deep learning components. Their sensitivity to environmental uncertainty and distribution shifts makes them difficult to verify formally with rigid assumptions [17]. Traditional assume-guarantee (A/G) reasoning can be applied, but a fundamental gap remains between A/G verification and the validity of those assumptions in a newly deployed environment [17].
A Probabilistic Framework for Verification and Validation
A unified probabilistic V&V methodology addresses this gap by combining frequentist-style verification with Bayesian-style validation, resulting in a flexible, nested guarantee for system safety [17].
The Unified V&V Workflow
The core methodology consists of three integrated steps: Abstraction, Verification, and Validation.
Diagram 1: Unified V&V workflow
Step 1: Abstraction: The concrete system ( ME ), including its neural perception components, is abstracted into a formal model that captures uncertainty. Using confidence intervals derived from data, an Interval Markov Decision Process (IMDP) abstraction ( \mathcal{M}E ) is constructed. This model over-approximates the behavior of ( M_E ) with a statistical confidence ( \alpha ), meaning it represents all possible systems consistent with the training environment ( E ) [17].
Step 2: Verification: A probabilistic model checker is used to verify a system-level temporal property ( \varphi ) (e.g., "the system remains safe") on the IMDP ( \mathcal{M}E ). The output is a verified upper bound ( \beta ) on the probability of the system violating ( \varphi ). Combined with Step 1, this yields a frequentist guarantee: "With confidence ( \alpha ), the system ( ME ) is safe with probability at least ( 1 - \beta )" [17].
Step 3: Validation: When deploying the model in a new environment ( E' ), this step quantifies how well the original abstraction ( \mathcal{M}E ) fits the new data. A Bayesian approach is used to construct a belief over the parameters of the new concrete model ( M{E'} ). "Intersecting" this belief with the IMDP's probability intervals produces a posterior probability ( 1 - \gamma ) that ( M{E'} ) is contained within ( \mathcal{M}E ) [17]. The final, nested guarantee is: "With confidence ( 1 - \gamma ), the deployed system ( M_{E'} ) satisfies the property ( \varphi ) with probability ( 1 - \beta )."
This framework elegantly handles the real-world challenge of evolving data distributions, providing a quantifiable and adaptable safety assurance rather than a brittle, absolute one.
Experimental Protocol for V&V of a Vision-Based System
This protocol outlines how to apply the unified V&V framework to a vision-based autonomous system, such as one used in a laboratory or manufacturing setting.
Objective: To verify the safety of a vision-based system under training conditions and validate the resulting guarantees against data from a deployment environment with potential distribution shifts.
Materials: See the "Research Reagent Solutions" table in Section 5.
Procedure:
Data Acquisition (Training Environment E):
- Collect ( N ) trajectories of the closed-loop system operating in the controlled training environment ( E ). Each trajectory should consist of time-synchronized ground truth states, images from the vision system, and applied control actions.
- Randomly hold out a portion (e.g., 20%) of this dataset for the initial abstraction step.
IMDP Abstraction:
- Perception Abstraction: Using the training data, compute confidence intervals for the perception system's state estimation error. For example, for each discrete state region, estimate the probability that the perceived state is within a bounded error of the ground truth. Use statistical techniques (e.g., conformal prediction) to derive intervals with confidence level ( \alpha ) [17].
- Dynamics Abstraction: Similarly, learn the confidence intervals for the system's dynamics transition probabilities between discrete states, accounting for both control actions and inherent stochasticity.
- Model Construction: Combine the perception and dynamics uncertainties into a single IMDP model ( \mathcal{M}_E ). This model's transition intervals encapsulate the combined uncertainty from both perception errors and stochastic dynamics.
Probabilistic Model Checking:
- Formally specify the safety property ( \varphi ) using temporal logic (e.g., "The system always avoids collision regions").
- Input the IMDP ( \mathcal{M}_E ) and property ( \varphi ) into a probabilistic model checker (e.g., PRISM, Storm).
- The tool will compute the maximum probability ( \beta ) of violating ( \varphi ) over all resolutions of the non-determinism in the IMDP. This is the verified safety bound.
Validation in Deployment Environment E':
- Deploy the trained model and collect a new dataset of ( M ) trajectories from the new environment ( E' ).
- Using Bayesian inference, update the belief over the parameters of the true system ( M_{E'} ) based on the new data.
- Calculate the posterior probability ( \gamma ) that ( M{E'} ) is *not* contained within the abstract model ( \mathcal{M}E ). This is the probability that the original model is invalid.
Reporting:
- The final output is the nested probabilistic guarantee: "With confidence ( 1 - \gamma ), the system ( M_{E'} ) is safe with probability at least ( 1 - \beta )."
- If ( \gamma ) is too high (e.g., > 0.05), the original model is likely invalid for ( E' ), indicating a significant distribution shift and triggering model adaptation or retraining.
Application Notes: Mitigation Strategies for Distribution Shifts
Once a distribution shift is detected and quantified, mitigation strategies are required. These can be implemented proactively during model development or reactively during deployment.
Table 2: Mitigation Strategies for Different Distribution Shifts
| Strategy | Mechanism | Applicable Shift Type | Implementation Consideration |
|---|---|---|---|
| Test-Time Refinement | Adjusts the model at inference time using an auxiliary objective or prior, without needing new labeled data from the new domain [75]. | Covariate, Concept | Low computational cost; ideal for foundation models. Improves OOD representation. |
| Distributionally Robust Optimization (DRO) | Trains models to perform well under a set of potential distribution shifts, often by optimizing for the worst-case scenario within a uncertainty set [76]. | All Types | Can lead to more conservative models; requires defining a realistic uncertainty set. |
| Domain Adaptation | Explicitly adapts a model trained on a source domain to perform well on a related but different target domain, often using unlabeled target data. | Covariate | Requires access to data from the target domain during training. |
| Continuous Monitoring & Performance Tracking | Establishes a framework for continuously monitoring model performance and key data distributions to detect deviations and trigger retraining [73] [74]. | All Types | Foundational practice for any deployed model; requires defining clear trigger thresholds. |
The following diagram illustrates the decision pathway for selecting and applying these strategies within a continuous lifecycle.
Diagram 2: Model monitoring and mitigation lifecycle
The Scientist's Toolkit: Research Reagent Solutions
The following table details key computational tools and methodologies essential for conducting the V&V and risk assessment protocols described in this document.
Table 3: Essential Research Reagents for AI/ML Model V&V
| Tool/Reagent | Function | Application Context |
|---|---|---|
| Interval MDPs (IMDPs) | A formal model that represents system uncertainty via intervals of transition probabilities, enabling rigorous abstraction of perception and dynamics [17]. | Core model for the unified V&V framework, used to capture the uncertainty from neural networks and environmental stochasticity. |
| Probabilistic Model Checkers (e.g., PRISM, Storm) | Software tools that automatically verify formal specifications (e.g., safety, liveness) against probabilistic models like IMDPs, providing mathematical guarantees [17]. | The "verification" engine in the V&V workflow; computes the safety probability bound ( \beta ). |
| Conformal Prediction | A statistical framework for producing prediction sets with guaranteed coverage levels under exchangeability, useful for quantifying uncertainty. | Can be used to derive the confidence intervals ( \alpha ) for the perception system's error in the abstraction step. |
| t-SNE / UMAP | Non-linear dimensionality reduction techniques for visualizing high-dimensional data in 2D or 3D, helping to identify clusters and distribution shifts [73]. | Exploratory data analysis to visually confirm covariate or label shifts between training and deployment datasets. |
| Fault Tree Analysis (FTA) | A top-down, deductive risk assessment method that identifies the potential causes of system failures [77]. | Used in system design to proactively identify how distribution shifts could lead to safety-critical failures. |
| Failure Modes and Effects Analysis (FMEA) | A proactive, systematic tool for identifying potential failure modes in a process or system and assessing their impact [77] [74]. | Applied during model development to prioritize risks associated with component failures under shift. |
Ensuring Robustness: Validation Frameworks and Comparative Analysis
Verification and Validation (V&V) are cornerstone processes for ensuring the reliability and correctness of complex computational models, particularly in safety-critical domains like autonomous systems and drug development. Verification addresses the question "Have we built the system correctly?" by checking whether a computational implementation conforms to its specifications. Validation answers "Have we built the right system?" by determining how accurately a model represents real-world phenomena. A probabilistic approach to V&V explicitly accounts for uncertainty, randomness, and distribution shifts, moving beyond binary guarantees to provide quantitative measures of confidence and reliability [17]. This framework is especially crucial for systems incorporating neural perception or other learning components, which are inherently stochastic and fragile to environmental changes [17].
Unifying verification with offline validation creates a powerful methodology for obtaining rigorous, flexible safety guarantees that can adapt to out-of-distribution test-time conditions [17]. This document provides detailed application notes and protocols for establishing such a framework, with a specific focus on probabilistic methods.
Core Concepts and Definitions
Foundational Principles of Probabilistic V&V
Probabilistic verification techniques comprise a rich set of formal methods for mathematically analyzing quantitative properties of systems or programs that exhibit stochastic, randomized, or uncertain behaviors [78]. Unlike classical verification, which asserts absolute correctness, probabilistic verification quantifies guarantees—such as almost-sure (probability 1) termination, bounded probability of failure, or expectation bounds—while managing both nondeterminism and probabilistic choice [78].
- Assume-Guarantee (A/G) Reasoning: A standard method for modular verification of complex systems [17]. In this framework, one specifies the assumptions on the system’s components, verifies each component's guarantees under these assumptions, and composes these to establish system-level properties [17].
- Perception Contracts: A specific application of A/G reasoning for vision-based systems that computes bounds on the accuracy of an observer (e.g., a neural network) sufficient to verify closed-loop system safety [17].
- Unified V&V Methodology: An approach that combines verification models of perception with their offline validation, leveraging models like interval Markov Decision Processes (IMDPs) to provide end-to-end guarantees adaptable to new environments [17].
Key Mathematical Frameworks
The algebraic and logical foundations of probabilistic verification build upon a generalization of standard logical frameworks. The weakest pre-expectation (wp) calculus is a fundamental tool, extending Dijkstra's concept from Boolean predicates to real-valued expectations [78]. This calculus systematically propagates quantitative postconditions (e.g., expected cost, probability of reaching an error state) backward through a program [78]. For a program command (C), state space (S), and post-expectation (\beta: S \rightarrow \mathbb{R}{\geq 0}), the corresponding wp transformer satisfies: [ \operatorname{wp}.C.\beta : S \rightarrow \mathbb{R}{\geq 0} ] with syntactic rules for commands like assignment, sequencing, probabilistic choice, and loops, the latter typically expressed as a least fixed point [78].
Table: Core Probabilistic Verification Techniques
| Technique | Core Function | Applicable Scope |
|---|---|---|
| Weakest Pre-expectation Calculus [78] | Propagates quantitative postconditions backward through programs. | Randomized algorithms; programs with probabilistic choice. |
| Interval Markov Decision Processes (IMDPs) [17] | Abstraction accounting for uncertainty with probability intervals. | Systems with perceptual uncertainty and environmental shifts. |
| Parametric Model Checking [78] | Replaces probabilities with symbolic parameters. | Software product lines; systems with configuration-dependent behavior. |
| Random Variable Abstraction (RVA) [78] | Abstracts state sets using linear functions and convex predicates. | Infinite-state probabilistic programs; automated invariant generation. |
Unified Probabilistic V&V Methodology
A proposed unified methodology for vision-based autonomy offers a concrete pathway for probabilistic V&V, consisting of three primary steps: abstraction, verification, and validation [17].
Methodology Workflow
The following diagram illustrates the integrated workflow of the unified probabilistic V&V methodology, showing the flow from system input to the final nested guarantee.
Detailed Workflow Description
- Abstraction Step: The neural perception's uncertainty is represented with confidence intervals, leading to an Interval Markov Decision Process (IMDP) abstraction, denoted (\mathcal{M}E) [17]. This abstraction overapproximates the concrete system (ME) with a specified confidence level (\alpha). In essence, (\mathcal{M}E) represents a set of possible closed-loop systems—accounting for uncertainties in the observer and dynamics—that are consistent with the data obtained from (ME) [17].
- Verification Step: A system-level temporal property (\varphi) (e.g., a safety requirement) is checked on the constructed IMDP (\mathcal{M}E) using a probabilistic model checker [17]. This step produces an upper bound (\beta) on the probability that a trajectory falsifies the property (\varphi) [17]. The combination of abstraction and verification yields a frequentist-style guarantee: "With confidence (1-\alpha) in the dataset, the chance that the underlying system (ME) produces a safe trajectory is at least (1-\beta)."
- Validation Step: The IMDP abstraction (\mathcal{M}E) is validated against a new deployment environment (E') [17]. This step adopts a Bayesian perspective to measure the probability that the new concrete model (M{E'}) is contained within (\mathcal{M}E). By constructing a belief over the parameters of (M{E'}) based on new data and "intersecting" it with the probability intervals in the IMDP, we compute a posterior probability (1-\gamma) that the new environment falls within the uncertainty of (\mathcal{M}E) [17]. This results in the final nested guarantee: "With confidence (1-\gamma), the system (M{E'}) satisfies the property (\varphi) with probability at least (1-\beta)."
Experimental Protocols and Application Notes
Protocol: Validation of a Vision-Based Autonomous System
This protocol details the application of the unified V&V methodology to a vision-based system, such as an autonomous vehicle or drone [17].
- Quick Definition: Validation of a vision-based autonomous system is a comprehensive procedure to verify the safety and reliability of a closed-loop system that uses deep neural networks for perception, providing probabilistic guarantees that are robust to environmental distribution shifts [17].
- Problem Statement: Deep vision models for processing rich sensory signals are fragile and vulnerable to distribution shifts, making it challenging to provide rigid, formal safety guarantees that hold in diverse deployment environments [17].
- Solution: Implement the three-step unified V&V methodology (abstraction, verification, validation) to obtain a flexible, nested probabilistic guarantee on system safety [17].
Implementation Steps:
Define Test Objectives and Specifications:
- Inputs: System requirements, identified safety-critical properties (\varphi) (e.g., "vehicle shall not collide"), training environment (E).
- Process: Formulate the temporal logic specification (\varphi) for verification. Define the confidence level (1-\alpha) for the abstraction. -Output: A formal, verifiable specification and a confidence target.
Execute Abstraction Step:
- Inputs: Collected execution traces (dataset (DE)) from the concrete system (ME) in the training environment.
- Process: Use statistical methods (e.g., conformal prediction, confidence interval estimation) on (DE) to build an IMDP abstraction (\mathcal{M}E) that overapproximates the system's behavior with confidence (1-\alpha) [17].
- Output: An IMDP model (\mathcal{M}_E).
Execute Verification Step:
Execute Validation Step for Deployment:
- Inputs: New dataset (D{E'}) from the deployment environment (E'), IMDP (\mathcal{M}E).
- Process:
- Compute a Bayesian posterior over the parameters of the true model (M{E'}) using (D{E'}) [17].
- Calculate the posterior probability (1-\gamma) that (M{E'}) is contained within the abstraction (\mathcal{M}E).
- Output: A model validity posterior (1-\gamma).
Report and Final Approval:
- Inputs: All previous outputs.
- Process: Synthesize results into the nested guarantee. Archive all documentation, models, and data for audit purposes.
- Output: A final validation report stating: "With confidence (1-\gamma), the system (M_{E'}) satisfies property (\varphi) with probability at least (1-\beta)."
- Expected Outcome: A validated vision-based autonomous system with a rigorous, flexible safety guarantee that adapts to new environments, enhancing trust and facilitating deployment [17].
Protocol: Formal Modeling and Verification of a Probabilistic Neural Network
This protocol outlines the formal verification of a probabilistic neural network model, such as one used for load balancing in a cloud environment [7].
- Quick Definition: Formal modeling and verification of a probabilistic neural network is a documented procedure that uses mathematical models and logical inference to ensure the correctness, reliability, and performance of a neural network model operating under uncertainty [7].
- Problem Statement: Machine learning models, including probabilistic neural networks (PNNs), are increasingly used for critical tasks like cloud load balancing, but they often lack formal verification, making it difficult to guarantee correctness and reliability in real-time systems [7].
- Solution: Develop a formal model of the PNN (e.g., an Effective Probabilistic Neural Network - EPNN) and verify its correctness using a formal method tool like Event-B and the Rodin platform [7].
Implementation Steps:
Define Test Objectives and Acceptance Criteria:
- Inputs: System requirements for the PNN (e.g., "EPNN shall select the best cluster for load distribution").
- Process: Define measurable acceptance criteria, such as probabilistic invariants (e.g., "the probability of selecting an overloaded cluster shall be less than 0.01") [7].
- Output: A set of verifiable properties and quantitative pass/fail thresholds.
Create Formal Model:
- Inputs: The PNN algorithm (e.g., EPNN architecture, Round Robin Assigning Algorithm).
- Process: Model the algorithm in the Event-B formal language. The model should include a context (static data) and a machine (dynamic behavior) [7].
- Output: An Event-B model of the PNN system.
Generate and Discharge Proof Obligations:
- Inputs: The Event-B model.
- Process: Use the Rodin tool to automatically generate proof obligations (theorems) that ensure the model's consistency and that its invariants are preserved. Discharge these proofs using automated and manual techniques [7].
- Output: A set of discharged proof obligations, confirming mathematical correctness.
Validate Model with Metrics:
- Inputs: The proven Event-B model, operational data or simulations.
- Process: Execute validation tests comparing the model's predicted behavior against real-world or simulated outcomes. Metrics may include system throughput, response time, and resource utilization [7].
- Output: Validation report with quantitative performance metrics.
Final Approval and Documentation Archival:
- Inputs: All proofs, validation results, and models.
- Process: Compile final report. Archive the entire development for future reference and regulatory audits.
- Output: A formally verified PNN model with documented evidence of correctness.
- Expected Outcome: A probabilistically sound neural network model with proven correctness and reliability, ensuring safe and predictable operation in critical computing environments [7].
The Scientist's Toolkit: Research Reagents and Solutions
This section details the essential computational tools and formal methods that constitute the "reagent solutions" for conducting probabilistic V&V research.
Table: Essential Research Reagents for Probabilistic V&V
| Item Name | Function | Application Context |
|---|---|---|
| Probabilistic Model Checker (e.g., PRISM, Storm) [78] | Automatically verifies probabilistic temporal properties against system models (MDPs, IMDPs). | Verification step; computing safety probability bounds (β). |
| Formal Modeling Tool (e.g., Event-B, Rodin Platform) [7] | Provides an environment for creating formal models and generating/discharging proof obligations. | Formal verification of algorithms and system models. |
| Weakest Pre-expectation (wp) Calculus [78] | A semantic framework for reasoning about expected values and probabilities in probabilistic programs. | Quantitative reasoning about randomized algorithms. |
| Interval Markov Decision Process (IMDP) [17] | A formal model that represents system uncertainty via intervals on transition probabilities. | Abstraction step; creating over-approximations of concrete systems. |
| Statistical Model Checker | Uses simulation and statistical inference to estimate the probability of system properties. | Approximate verification of complex systems where exhaustive model checking is infeasible. |
| Bayesian Inference Engine | Updates beliefs about model parameters based on observed data from new environments. | Validation step; computing the model validity posterior (γ). |
Best Practices for Protocol Implementation
Implementing a robust probabilistic V&V framework requires adherence to several key best practices, drawn from both software engineering and formal methods.
Established Best Practices
- Establish Risk-Based Testing Approaches: Design validation protocols that prioritize testing efforts based on risk assessment. Focus more resources on high-risk areas (e.g., safety-critical components) while maintaining appropriate coverage for lower-risk elements [79]. Do: Conduct thorough risk assessments to identify critical functions, analyze potential failure modes, and allocate testing resources proportionally to risk levels. Don't: Apply uniform testing intensity across all areas regardless of risk [79].
- Maintain Clear Traceability Throughout: Ensure every validation protocol element can be traced back to specific requirements and forward to test results, creating comprehensive audit trails for regulatory compliance [79]. Do: Create detailed traceability matrices linking requirements to test cases. Maintain unique identifiers for all protocol elements and document all changes with proper justification [79].
- Define Measurable Acceptance Criteria: Establish specific, quantifiable acceptance criteria that eliminate subjective interpretation and provide clear pass/fail determinations for all validation activities [79]. Do: Use numerical thresholds, specific performance metrics, and objective evaluation methods with clear measurement procedures and tolerance limits. Don't: Use vague terms like 'acceptable' or 'satisfactory' without quantification [79].
- Implement Robust Change Control: Establish formal procedures for managing protocol changes that maintain validation integrity while allowing necessary updates [79]. Do: Document all protocol changes with impact assessments, obtain appropriate approvals before implementation, and maintain version control with clear change histories [79].
- Plan for Protocol Reusability: Design validation protocols as reusable templates that can be adapted for similar projects while maintaining consistency and reducing development time [79]. Do: Create modular protocol structures with standardized sections and develop template libraries for common validation scenarios [79].
Visualization of the Probabilistic V&V Ecosystem
The following diagram maps the logical relationships between the key components, tools, and outputs of the probabilistic V&V ecosystem, illustrating how they interconnect to form a cohesive framework.
The adoption of probabilistic modeling has become a cornerstone of modern scientific research, particularly in fields characterized by high stakes and inherent uncertainty, such as drug development and clinical medicine. These approaches provide a structured framework to quantify uncertainty, integrate diverse data sources, and support complex decision-making processes. The core of this paradigm lies in its ability to distinguish between different types of uncertainty—epistemic (arising from incomplete knowledge) and aleatoric (stemming from natural variability)—allowing for more nuanced predictions and risk assessments [80]. Within the context of model verification and validation (V&V) research, probabilistic methods are not merely computational tools; they are essential components for building credible, trustworthy, and clinically actionable digital twins and predictive models. This analysis examines the trade-offs between prominent probabilistic modeling frameworks, focusing on their application to specific use cases in precision medicine and pharmaceutical development, and provides detailed protocols for their implementation.
Comparative Analysis of Probabilistic Modeling Frameworks
Different probabilistic modeling approaches offer distinct advantages and limitations, making them suited to particular problems. The table below summarizes the key characteristics of several prominent frameworks.
Table 1: Comparative Analysis of Probabilistic Modeling Approaches
| Modeling Approach | Primary Use Case | Key Strengths | Key Limitations | Verification & Validation Considerations |
|---|---|---|---|---|
| Bayesian Networks (BNs) & Influence Diagrams [81] | Clinical trial design with competing outcomes; Integrating clinical expert knowledge. | - Incorporates competing factors & expert judgment via prior distributions.- Intuitive visual representation of variable dependencies. | - Model structure can be complex to define.- Requires careful specification of prior distributions. | - Verification involves checking the consistency of the conditional probability tables.- Validation requires assessing predictive accuracy against held-out clinical data. |
| Markov Models [81] | Modeling disease progression over time; Health economic evaluations (e.g., QALY calculation). | - Excellently handles time-dependent processes and transitions between health states.- Well-established for calculating long-term outcomes. | - Can suffer from the "memoryless" property assumption.- State space can become large and computationally expensive. | - Verification involves ensuring state transition probabilities sum to one.- Validation involves comparing predicted disease trajectories to real-world longitudinal patient data. |
| Probabilistic Graphical Regression (e.g., GraphR) [82] | Genomic network inference; Modeling sample heterogeneity (e.g., inter-tumoral variations). | - Explicitly accounts for sample-level heterogeneity, avoiding biased estimates.- Infers sparse, sample-specific networks. | - Computationally intensive for very high-dimensional data.- Requires careful tuning of sparsity parameters. | - Verified via simulation studies assessing network structure recovery.- Validated by its ability to reveal biologically meaningful, heterogeneous network structures missed by homogeneous models. |
| Formally Verified Probabilistic Neural Networks (PNNs) [7] [17] | Safety-critical autonomous systems; Real-time cloud load balancing. | - Combines predictive adaptability with mathematical guarantees of correctness.- Suitable for dynamic, uncertain environments. | - Formal verification process can be complex and requires specialized expertise.- Limited application in clinical domains to date. | - Verification uses tools like Event-B and Rodin for automated and manual proof generation [7].- Validation involves rigorous testing in domain-shifted environments to ensure robustness [17]. |
| Digital Twins with VVUQ [80] | Personalized patient treatment simulation; Predicting health trajectories under interventions. | - Provides a dynamic, continuously updated virtual representation of an individual.- Mechanistic models support causal inference. | - Requires high-frequency, high-quality real-time data from biosensors.- VVUQ process is complex and must be iterative. | - Involves rigorous Verification, Validation, and Uncertainty Quantification (VVUQ).- Validation is ongoing ("temporal validation") as the twin updates with new patient data [80]. |
Application Notes & Experimental Protocols
Application Note 1: Clinical Trial Design as a Decision Problem
1.1 Background and Objective Traditional clinical trial design often focuses on a single primary outcome with statistical power calculations. Viewing a trial as a formal decision problem allows for the incorporation of competing outcomes, such as efficacy versus toxicity, and the integration of patient heterogeneity [81]. The objective is to design a trial that maximizes a composite utility function, such as Quality-Adjusted Life Years (QALYs), rather than merely testing a statistical hypothesis.
1.2 Workflow and Signaling Pathway The following diagram illustrates the integrated workflow for a probabilistic clinical trial design, combining a Bayesian Network for initial probability assessment with a Markov Model for long-term outcome projection.
1.3 Detailed Experimental Protocol
Protocol Title: Probabilistic Design of a Non-Inferiority Trial for HPV+ Oropharyngeal Cancer Using QALY Maximization.
Objective: To determine the optimal radiation dose (70 Gy vs. 55 Gy) by evaluating the trade-off between tumor control probability (TCP) and normal tissue complication probability (NTCP) using a QALY-based utility function.
Materials and Reagents: Table 2: Research Reagent Solutions for Clinical Trial Modeling
Reagent / Tool Function / Explanation Influence Diagram A Bayesian network incorporating decision nodes (e.g., dose) and a utility node (QALY). It visually structures the decision problem [81]. Tumor Control Probability (TCP) Model A logistic function (e.g., ( TCP(D) = 1 / (1 + (D50/D)^{γ50}) )) that estimates the probability of tumor control as a function of radiation dose D [81]. Normal Tissue Complication Probability (NTCP) Model A logistic function estimating the probability of complications (e.g., xerostomia) for a given dose distribution to an organ-at-risk [81]. Markov Model States Defined health states (e.g., "Post-RT," "Recurrent Disease," "Gr2+ Xerostomia," "Death") through which a simulated patient cohort transitions annually [81]. Monte Carlo Simulation Engine Software that performs random sampling from the parameter distributions (e.g., D50, γ50) to propagate uncertainty and compute a distribution of expected QALYs [81]. Methodology:
- Model Structuring: Construct an influence diagram with expert clinical input. Key chance nodes include patient risk factors (HPV status, smoking history), TCP, and NTCP. The decision node is radiation dose, and the utility node is QALY [81].
- Parameter Estimation:
- TCP: Define prior distributions for parameters D50 (dose for 50% control) and γ50 (slope). Using historical data (e.g., 82.4% 3-year survival at 70 Gy), calibrate the model so that the survival difference at 55 Gy reflects scenarios of interest (e.g., 7%, 10%, 15% reduction). The Monte Carlo process will sample from these parameter distributions [81].
- NTCP: Fit logistic NTCP models for complications like xerostomia using clinical dose-volume histogram data and toxicity outcomes.
- Markov Model Setup: Define health states, annual transition probabilities between states (informed by the BN output and literature), and assign utility weights (0-1) to each state. "Death" absorbs all patients and has a utility of 0.
- Simulation and Analysis: Run a Monte Carlo simulation (e.g., 10,000 iterations) for each dose strategy. For each iteration, sample parameters from the priors, calculate initial state probabilities from the BN, run the Markov cohort simulation for a defined time horizon (e.g., 20 years), and accumulate QALYs.
- Decision Rule: The optimal dose is the one with the highest mean expected QALY across all Monte Carlo iterations. Conduct probabilistic sensitivity analysis on key parameters to test the robustness of the decision.
Application Note 2: Calculating Probability of Success for Drug Development
2.1 Background and Objective The decision to progress a drug candidate from Phase II to Phase III trials is a critical, high-risk milestone. Probability of Success (PoS) provides a quantitative framework to support this "go/no-go" decision by quantifying the uncertainty of achieving efficacy in a confirmatory trial [83]. The objective is to calculate a PoS that robustly integrates all available data, including Phase II results and external information, to minimize attrition in late-stage development.
2.2 Workflow for Probability of Success Calculation The diagram below outlines the key steps and data sources involved in a comprehensive PoS calculation.
2.3 Detailed Experimental Protocol
Protocol Title: Bayesian Calculation of Probability of Success for a Phase III Trial Using Integrated Phase II and Real-World Data.
Objective: To compute the probability that a Phase III trial will demonstrate a statistically significant and clinically meaningful effect on the primary efficacy endpoint, leveraging Phase II data and external real-world data (RWD) to construct an informative design prior.
Materials and Reagents: Table 3: Research Reagent Solutions for PoS Calculation
Reagent / Tool Function / Explanation Design Prior A probability distribution (e.g., Normal) representing the current uncertainty about the true treatment effect size in the Phase III population. It is the cornerstone of the PoS calculation [83]. Real-World Data (RWD) Data from patient registries, electronic health records, or historical clinical trials. Used to inform the design prior, especially when Phase II uses a surrogate endpoint [83]. Predictive Power / Assurance The statistical method for calculating PoS. It is the expected power of the Phase III trial, averaged over the possible effect sizes described by the design prior [83]. Computational Software (e.g., R, Stan) Environment for performing Bayesian analysis and Monte Carlo simulations to compute the PoS. Methodology:
- Endpoint Mapping: If the Phase II endpoint is a biomarker or surrogate, establish its relationship with the Phase III clinical endpoint using historical data or RWD. This model will be used to translate Phase II results into a prior for the clinical endpoint.
- Construct the Design Prior:
- Option A (Phase II data on clinical endpoint): Use the Phase II results (posterior distribution of the effect) as the prior for the Phase III effect.
- Option B (Phase II data on surrogate endpoint): Use the established relationship from Step 1 to convert the Phase II surrogate effect into a predictive distribution for the Phase III clinical effect.
- Incorporate RWD: Use RWD to refine the prior, for example, by providing a more precise estimate of the control group response rate or the association between surrogate and clinical endpoints.
- Define Phase III Trial Design: Specify the planned sample size, randomization ratio, primary endpoint, and statistical test (e.g., one-sided log-rank test at α=0.025).
- Calculate Probability of Success:
- For a range of possible treatment effects ( θ ), sample from the design prior.
- For each sampled ( θ ), calculate the power of the Phase III design to detect that effect.
- The PoS is the average of these power values across all samples from the prior. This can be computed via numerical integration or Monte Carlo simulation.
- Sensitivity and Scenario Analysis: Test the robustness of the PoS by varying the design prior (e.g., using a more skeptical or optimistic prior) and the Phase III trial assumptions (e.g., sample size).
The selection of a probabilistic modeling approach is a critical decision that must be guided by the specific use case, the nature of the available data, and the required level of verification and validation. As demonstrated, Bayesian networks and Markov models are powerful for structuring complex clinical decisions with multiple competing outcomes, while Probability of Success frameworks are indispensable for de-risking pharmaceutical development. Emerging paradigms, such as formally verified neural networks and digital twins subject to rigorous VVUQ, represent the frontier of probabilistic modeling, offering the potential for adaptive, personalized, and provably reliable systems. The ongoing challenge for researchers and practitioners is to continue refining these methodologies, standardizing VVUQ processes, and fostering interdisciplinary collaboration to ensure that probabilistic models can be trusted to inform high-stakes decisions in medicine and beyond.
In the domain of probabilistic model verification and validation (V&V), a significant challenge is maintaining model reliability in the face of evolving real-world conditions. Cross-sectional data analysis, which involves examining a dataset at a fixed point in time, provides a powerful foundation for building dynamic validation frameworks [84] [85]. This approach enables researchers to create "snapshots" of system performance, which can be sequentially compared to detect performance degradation and trigger model updates [86].
Calendar period estimation extends this capability by analyzing cross-sectional data collected across multiple time points, allowing for the detection of temporal trends and the calendar-year impacts of policy changes or external factors [87] [88]. Within drug development, this methodology has demonstrated particular value for estimating the conditional probability of drugs transitioning through clinical trial phases, providing a more timely alternative to longitudinal cohort studies [87]. This Application Note details the protocols for implementing these techniques to achieve robust, dynamic validation of probabilistic models.
Theoretical Foundation
Cross-Sectional Data in Dynamic Analysis
Cross-sectional data analysis involves observing multiple variables at a single point in time, providing a static snapshot of a population or system [84] [85]. In dynamic validation, sequential cross-sectional snapshots are compared to infer temporal changes, overcoming the limitation of single-time-point analysis.
- Definition and Characteristics: A cross-sectional dataset records observations of multiple variables for multiple economic entities (e.g., drugs in development, autonomous systems) at a particular time
t[84]. The sequence of data recording is irrelevant in a single cross-section, unlike time-series data where sequence is meaningful [85]. - Advantages for Dynamic Systems: When collected repeatedly, cross-sectional data enables a "quasi-experimental design" [85]. This facilitates monitoring the effects of societal or secular change on a population's characteristics, providing "net effects" crucial for detecting model drift and performance degradation.
Calendar Period Estimation
Calendar period estimation is a specific application of repeated cross-sectional analysis that measures how transition probabilities or system behaviors change across calendar time.
- Core Principle: This method constructs a series of "current transition tables" as calendar time progresses, rather than following a fixed cohort over time (a longitudinal design) [88]. This allows for a more dynamic understanding of processes as they unfold.
- Application in Drug Development: Traditional longitudinal studies require observing drug cohorts for over a decade to estimate success probabilities, creating significant delays in detecting trends [88]. The cross-sectional, calendar period approach enables timely detection and measurement of the impact of policy changes, scientific advancements, or operational practices on transition probabilities [87] [88].
Integration with Probabilistic V&V
The unification of cross-sectional data with probabilistic verification creates a nested guarantee framework. A model M_E is verified in a source environment E to provide a probabilistic safety guarantee, contingent on the model's validity [17]. Cross-sectional data from a new deployment environment E' is then used to validate this assumption, producing a quantitative posterior chance (1-γ) that the new environment falls within the model's uncertainty bounds [17]. This yields a final nested guarantee: with confidence 1-γ, the system in E' satisfies the safety property with probability 1-β.
Application Notes & Protocols
Protocol 1: Constructing a Calendar Period Estimation Pipeline
This protocol outlines the process for building a dynamic validation system using cross-sectional data to estimate transition probabilities over time, adaptable to both clinical drug development and autonomous system testing.
Objective: To establish a proactive/reactive pipeline for tracking system state transitions and dynamically updating performance models.
Diagram 1: Dynamic updating pipeline workflow.
- Step 1: System State Definition: Define all discrete states or phases in the process. In drug development, these are typically Phase I, Phase II, Phase III, and Approval [88]. For autonomous systems, states could be Operational Design Domain (ODD) A, ODD B, System Failure, etc.
- Step 2: Cross-Sectional Data Collection: At regular intervals (e.g., monthly, quarterly), collect snapshots of all entities in the system and their current states. The data should include key characteristics (e.g., drug modality, therapeutic area [87]; for autonomous systems, software version, sensor suite).
- Step 3: Life Table Construction: For each calendar period (e.g., yearly from 2002-2022 [87]), and for each state/phase, construct a life table. This table tracks the number of entities entering the state, the number remaining at each time interval, and the number transitioning out.
- Step 4: Hazard Graduation: The raw hazard of transition from the life tables can be noisy. Graduate these hazards using a Generalized Additive Model (GAM) to produce a smooth hazard function over time [87]. This accounts for non-linear trends.
- Step 5: Probability Calculation: From the smoothed hazards, calculate two key metrics:
- Conditional Probability of Transition: The probability of leaving a state given the time already spent in that state.
- Overall Probability of Success (PoS): The cumulative probability of transitioning from an initial state (e.g., Phase I) to a successful terminal state (e.g., Approval), computed as the product of the conditional transition probabilities across all phases.
- Step 6: Update Decision (Proactive vs. Reactive):
- Proactive Updating: A candidate updated model is tested every time new cross-sectional data becomes available [86].
- Reactive Updating: An update is only triggered when the performance of the current model degrades below a pre-set threshold or when a significant change in the system structure is detected [86].
Protocol 2: Dynamic Validation with Interval Markov Models
This protocol uses cross-sectional data to build and validate Interval Markov Decision Process (IMDP) abstractions, providing robust safety guarantees for systems with perceptual uncertainty, such as vision-based autonomy.
Objective: To create a unified verification and validation (V&V) framework that provides safety guarantees adaptable to distribution shifts in the deployment environment.
Diagram 2: IMDP abstraction and validation process.
Step 1: Abstraction (Building the IMDP):
- Collect cross-sectional data from the source environment
E(e.g., a controlled test track for a robot). - Represent uncertainties from neural perception and system dynamics using confidence intervals. This leads to an Interval Markov Decision Process (IMDP) abstraction,
ℳ_E[17]. - The IMDP
ℳ_Eover-approximates the concrete systemM_Ewith a statistical confidence(1-α), meaning it contains the behavior distribution ofM_Ewith high probability [17].
- Collect cross-sectional data from the source environment
Step 2: Verification:
- Define a system-level temporal safety property
φ(e.g., "the robot never collides"). - Use a probabilistic model checker (e.g., PRISM [17]) to verify
φon the IMDPℳ_E. - The model checker produces an upper bound
βon the probability of violatingφ[17]. This yields a frequentist-style guarantee: "With confidence(1-α)in the dataset, the chance that the underlying systemM_Eis safe is at least(1-β)."
- Define a system-level temporal safety property
Step 3: Validation in a New Environment:
- Deploy the system in a new environment
E'and collect a fresh set of cross-sectional data. - Instead of a frequentist hypothesis test, adopt a Bayesian perspective. Construct a belief (posterior distribution) on the parameters of the new concrete model
M_{E'}based on the new data [17]. - "Intersect" this belief with the probability intervals in the original IMDP
ℳ_Eto compute a quantitative posterior probability(1-γ)thatM_{E'}is contained withinℳ_E[17].
- Deploy the system in a new environment
Step 4: Final Nested Guarantee:
- The output is a nested probabilistic guarantee: "With confidence
(1-γ), the systemM_{E'}in the new environment satisfies the safety propertyφwith probability at least(1-β)" [17]. - This guarantee elegantly combines the rigor of formal verification (frequentist) with the flexibility of empirical validation (Bayesian), allowing safety claims to adapt to distribution shifts.
- The output is a nested probabilistic guarantee: "With confidence
The following tables summarize key quantitative findings from the application of these methods in different domains.
Table 1: Drug Development Transition Probability Analysis (Based on [87] [88])
| Characteristic | Impact on Transition Probabilities | Data Source & Method |
|---|---|---|
| Therapeutic Indication | Transition propensity and overall Probability of Success (PoS) vary significantly by disease area. | Pharmaprojects database; Life tables & GAM smoothing for 2002-2022. |
| Mechanism of Action | Heavily influences the likelihood of transitioning out of each clinical phase. | Pharmaprojects database; Calendar period estimation. |
| Temporal Trends | Positive trends in overall PoS for certain drug classes, suggesting improving industry productivity. | Cross-sectional analysis over calendar time (2002-2022). |
| Overall Attrition | Only ~10% of candidates entering human trials achieve FDA approval. Primary causes: Lack of efficacy (40-50%), toxicity (30%). | Historical cohort analysis embedded in cross-sectional data. |
Table 2: Performance of Dynamic Updating Pipelines in Clinical Prediction (Based on [86])
| Updating Pipeline Type | Performance Outcome | Context |
|---|---|---|
| Proactive Updating | Better calibration and discrimination than a non-updated model. | Cystic Fibrosis 5-year survival prediction model over a 10-year dynamic updating period. |
| Reactive Updating | Better calibration and discrimination than a non-updated model. | Cystic Fibrosis 5-year survival prediction model over a 10-year dynamic updating period. |
| No Updating (Baseline) | Performance degradation over time. | Used as a comparator for the proactive and reactive pipelines. |
The Scientist's Toolkit
Table 3: Essential Research Reagent Solutions
| Item | Function & Application |
|---|---|
| Pharmaprojects (Citeline) Database | A global database tracking drug candidates from pre-clinical stages to market launch or discontinuation. Provides the foundational cross-sectional data on drug development timelines and outcomes. [87] [88] |
| Probabilistic Model Checker (e.g., PRISM) | A software tool for formally verifying probabilistic models like IMDPs against temporal logic specifications. Used to compute the upper bound β on the probability of safety property violation. [17] |
| Generalized Additive Model (GAM) | A statistical modeling technique used to graduate (smooth) raw hazard functions estimated from life tables. Captures complex, non-linear trends in transition probabilities over time. [87] |
| Interval Markov Decision Process (IMDP) Framework | A formal modeling abstraction that represents system uncertainties as intervals of transition probabilities. Serves as the core mathematical structure for unifying verification and validation under uncertainty. [17] |
| Event-B Formal Modeling Tool (Rodin Platform) | A formal method toolset for system-level modeling and verification. Used for proving the correctness of algorithms, such as load-balancing mechanisms in probabilistic neural networks. [7] |
Within the framework of probabilistic approaches to model verification and validation, benchmarking against real-world clinical transition probabilities represents a critical methodology. This process assesses the predictive accuracy of computational models in healthcare by comparing their outputs to empirically observed probabilities of patient health state transitions [24]. Such benchmarking is fundamental for transforming models from theoretical constructs into trusted tools for drug development and clinical decision-making. The core challenge lies in reconciling model predictions with real-world evidence, a task that requires sophisticated statistical techniques to account for inherent uncertainties—both aleatory (natural variability in patient outcomes) and epistemic (uncertainty due to limited data) [24].
The integration of generative artificial intelligence (AI) offers a paradigm shift in this domain. By processing patient narratives and free-text outcomes in a flexible, context-aware manner, generative AI supports a bottom-up, narrative-based approach to understanding health experiences [89]. This stands in contrast to traditional, reductionist methods that often struggle to capture the multidimensional nature of lived health. Furthermore, probabilistic model checking provides a formal verification technique for stochastic systems, using temporal logic to algorithmically check properties against a model. This approach is increasingly valuable for quantifying the reliability and timeliness of outcomes in the context of uncertain clinical trajectories [25].
Theoretical Foundations and Data Presentation
Core Concepts in Probabilistic Validation
Model validation in this context is defined as the process of assessing a model's predictive capability against experimental data, while acknowledging that all models are approximations of their target phenomena [24]. A functional analytic, probabilistic framework is particularly well-suited for this task, as it can represent random quantities using expansions like Polynomial Chaos Expansions (PCEs) to characterize uncertainty [24]. The comparison between computation and observation is formalized through a validation metric, which quantifies the agreement for a specific Quantity of Interest (QOI), such as the probability of a clinical event within a defined timeframe [24].
Clinical Transition Probability Data
The following tables summarize quantitative data relevant for benchmarking models of patient state transitions in specific therapeutic areas.
Table 1: Example Transition Probabilities in Rheumatology and Oncology Clinical Trials
| Therapeutic Area | Clinical State Transition | Probability Range | Key Influencing Factors | Data Source |
|---|---|---|---|---|
| Rheumatology | Improvement in patient-reported pain scores | Subject to model calibration | Disease activity, treatment regimen | Clinical trial workflow ethnography [90] |
| Oncology | Tumor response to therapy | Subject to model calibration | Cancer stage, biomarker status | Clinical trial workflow ethnography [90] |
| General | Maximum acceleration in a dynamical system exceeding a threshold | Validated via statistical hypothesis test | System parameters, shock load | Model validation challenge exercise [24] |
Table 2: Time-Motion Analysis of Clinical Trial Workflow Activities
| Staff Role | Activity Duration Profile | Frequency of Activities | Common Workflow Bottlenecks |
|---|---|---|---|
| Nurses | Highest total time consumption | High frequency of short-duration tasks | Tasks requiring full commitment of CRCs [90] |
| Clinical Research Coordinators (CRCs) | Variable | Managing 5-6 trials concurrently | Transferring notes from paper to computers [90] |
| Administrative Assistants | More activities at workflow start/end | Moderate frequency | Deviations from Standard Operating Procedures (SOPs) [90] |
Experimental Protocols
Protocol 1: Probabilistic Model Validation for Clinical Transition Models
This protocol outlines a structured procedure for validating computational models that predict clinical transition probabilities [24].
1. Objective: To assess the predictive accuracy of a clinical transition probability model by comparing its outputs to real-world observational or trial data.
2. Materials and Reagents:
- Computational Model: The candidate model encoding hypothesized transition dynamics.
- Validation Dataset: A high-quality, independent dataset of observed patient state transitions.
- Uncertainty Quantification (UQ) Software: Tools like PRISM [25] or Storm [25] for probabilistic analysis.
- Statistical Computing Environment: Software (e.g., R, Python) for performing calibration and hypothesis tests.
3. Procedure:
- Step 1: Define Quantity of Interest (QOI): Pre-specify the clinical transition to be validated (e.g., "probability of disease progression within 12 months").
- Step 2: Model Calibration: Use a statistical procedure (e.g., maximum likelihood estimation) to calibrate the model's uncertain parameters against a subset of the available experimental data. Represent the inherent uncertainty of these parameters using Polynomial Chaos Expansions (PCEs) [24].
- Step 3: Uncertainty Propagation: Propagate the characterized parameter uncertainties through the computational model to obtain a probabilistic prediction for the QOI. The stochastic Galerkin method is well-suited for this when using PCEs [24].
- Step 4: Compute Validation Metric: Compare the model's probabilistic prediction (from Step 3) against the real-world outcomes in the validation dataset. Implement a simple statistical hypothesis test to quantify the agreement (e.g., testing if the observed data falls within the model's prediction intervals) [24].
- Step 5: Decision for Validation: Based on the validation metric, assess whether the model's predictive capability is sufficient for its intended use context.
Protocol 2: Workflow Ethnography for Identifying Real-World Transition Probabilities
This protocol describes how to collect real-world data on clinical processes that can inform transition probabilities, particularly those related to operational and patient-reported outcomes [90].
1. Objective: To model the operational workflow of clinical trials and identify bottlenecks and time-motion data that can impact the measurement of clinical transitions.
2. Materials and Reagents:
- Ethnographic Field Notes: Tools for qualitative data recording.
- Time-Motion Database: A structured database (e.g., MySQL) with a web browser interface for real-time data entry [90].
- UML Modeling Software: An environment like Eclipse with a UML2 plugin for standardizing workflow diagrams [90].
3. Procedure:
- Step 1: Site Observation and Interviewing: Conduct ethnographic observations at clinical trial sites. Interview key personnel (CRCs, Principal Investigators) to understand workflow, variations, and perceived points of failure [90].
- Step 2: Hierarchical Activity Classification: Transcribe field notes and compile a list of activities. Classify them hierarchically into major and minor activities through researcher consensus [90].
- Step 3: Time-Motion Data Entry: For each task, record the start and end times to the nearest second in the dedicated database. When staff engage in concurrent tasks, rank them as primary and secondary activities [90].
- Step 4: Workflow Modeling in UML: Model the observed workflow using Unified Modeling Language (UML) Activity Diagrams. Represent events as "Opaque Actions" connected by "Control Flows," and use "Decision Nodes" for workflow forks [90].
- Step 5: Standardization via UML Profile: Apply a developed UML profile to the workflow model. This profile standardizes the representation by incorporating data tags for time, distribution, and efficiency, enabling comparative analysis across international sites [90].
Visualization of Methodologies
Probabilistic Model Validation Workflow
The following diagram illustrates the integrated workflow for validating a model of clinical transition probabilities, from data preparation to the final validation decision.
Clinical Trial Workflow Ethnography
This diagram outlines the protocol for conducting ethnographic studies to capture real-world clinical processes and their associated transition probabilities.
The Scientist's Toolkit
Table 3: Key Research Reagent Solutions for Probabilistic Clinical Model Validation
| Item Name | Function/Brief Explanation |
|---|---|
| Polynomial Chaos Expansions (PCEs) | A functional analytic method to represent uncertain model parameters as generalized Fourier expansions, facilitating uncertainty quantification and propagation [24]. |
| Probabilistic Model Checkers (PRISM, Storm) | Software tools that perform formal verification of stochastic systems against temporal logic properties, checking measures like "probability of a clinical event within a time bound" [25]. |
| Unified Modeling Language (UML) Profile | A standardized extension to UML for clinical trial workflows, enabling consistent representation and comparative analysis of operational processes across different research sites [90]. |
| Generative AI / Large Language Models (LLMs) | AI tools capable of processing patient narratives and free-text outcomes to generate qualitative insights and synthesize patient experiences, moving beyond reductionist scores [89]. |
| Stochastic Galerkin Method | A numerical technique for propagating uncertainty through a computational model, particularly efficient when model parameters are represented using PCEs [24]. |
In the domain of mission-critical systems, from aerospace to cloud computing, ensuring algorithmic correctness is not merely a best practice but a fundamental requirement. Formal methods provide a mathematical basis for specifying and verifying that systems behave as intended. Event-B is a prominent formal method for system-level modelling and analysis, utilizing set theory as its modelling notation and refinement to represent systems at different abstraction levels [91]. Its core strength lies in using mathematical proof to verify consistency between these refinement levels, thereby providing a rigorous framework for demonstrating algorithmic correctness [92]. This application note details how Event-B and its supporting tools can be employed within a broader probabilistic approach to model verification and validation (V&V) research, offering researchers a pathway to high-assurance system development.
The Rodin Platform, an Eclipse-based IDE for Event-B, provides effective support for refinement and mathematical proof [91]. As an open-source tool, it forms a cornerstone for practical formal verification projects. The platform's extendable nature, through plugins, allows its capabilities to be tailored to specific research needs, such as incorporating probabilistic reasoning [91] [93].
Event-B Methodology and Toolchain
Core Principles of the B-Method and Event-B
The B-Method, and its evolution Event-B, is a formal method based on an abstract machine notation [92]. The development process follows a structured approach:
- Abstract Machine Specification: The initial, most abstract model specifies the system's goal and core properties using set theory and first-order logic, focusing on what the system should do, not how [92].
- Refinement: Through a series of refinement steps, details about data structures and algorithms are progressively added to the abstract model. Each refinement step must be proven to be coherent and preserve all properties of the more abstract machine [92].
- Implementation: The refinement process continues until a deterministic version is achieved, which can be translated into executable programming language code [92].
A key feature of this method is the use of the same notation throughout the entire development cycle, from specification to implementation, reducing the potential for errors introduced during translation [92].
The Verification Toolchain Ecosystem
Several robust tools support the B-Method and Event-B, facilitating industrial application:
- Rodin Platform: The primary open-source tool for Event-B, supporting refinement and mathematical proof within an Eclipse IDE. It is extendable with plugins [91] [92].
- Atelier B: An industrial-strength tool for the classical B-Method, renowned for its use in developing safety-critical software for systems like the automatic Paris Métro lines and the Ariane 5 rocket [92].
- B-Toolkit: A collection of tools, including a mathematical interpreter based on set theory, designed to support the B-Method [92].
- Event-B Qualitative Probability Plug-in: A specific Rodin plugin that supports reasoning about almost-certain termination, a key property for probabilistic algorithms [93].
Table: Key Tools for the B-Method and Event-B
| Tool Name | Method | License Model | Primary Use Cases |
|---|---|---|---|
| Rodin Platform | Event-B | Open Source | Academic research, system-level modelling and analysis |
| Atelier B | B-Method | Commercial (Community Edition available) | Industrial safety-critical software (transport, aerospace) |
| B-Toolkit | B-Method | Source Available | Formal development and verification |
| Qualitative Probability Plug-in | Event-B | Open Source | Reasoning about probabilistic convergence |
Protocol for Probabilistic Verification in Event-B
This protocol outlines the steps for modelling and verifying a probabilistic system, specifically focusing on proving almost-certain termination using the Qualitative Probability plug-in [93].
Experimental Setup and Research Reagent Solutions
Table: Research Reagent Solutions for Event-B Probabilistic Modelling
| Item | Function | Example/Description |
|---|---|---|
| Rodin Platform | Core IDE | Eclipse-based environment for Event-B model development, refinement, and proof [91]. |
| Qualitative Probability Plug-in | Adds probabilistic reasoning | Enables marking events as probabilistic and proving almost-certain termination [93]. |
| Model Context | Defines static structures | Contains sets, constants, and axioms that form the mathematical basis of the model. |
| Model Machine | Defines dynamic behavior | Contains variables, invariants, and events that model the system's state transitions. |
| Proof Obligations | Verification artifacts | Mathematical formulas generated by Rodin to verify model consistency and refinement. |
Detailed Methodology
The following workflow diagrams the process of creating and verifying a probabilistic model in Event-B, using the example of a contention resolution protocol [93].
Step 1: Develop a Non-Probabilistic Abstract Model. Begin by modelling the system's core functionality without probabilistic details. For the contention resolution example, this involves defining variables (e.g., p1_choice and p2_choice to represent choices of two processes) and an event (e.g., resolve) with a guard that triggers when choices are identical [93].
Step 2: Introduce Probabilistic Convergence. Identify the event that should lead to termination with probability 1. Using the Qualitative Probability plug-in:
- Set the event's convergence attribute to
convergent. - Change the event's probabilistic attribute from
standardtoprobabilistic[93].
Step 3: Define Variant and Bound. A variant is an expression that must be shown to probabilistically decrease.
- Define a Variant: Introduce a numeric expression (e.g.,
VARIANT = 1in simple cases) that is part of the model's state. - Add a Bound Element: Create a bound element (a special construct in the plug-in) to specify an upper limit for the variant, ensuring it does not increase indefinitely [93].
Step 4: Generate and Discharge Proof Obligations (POs). The Rodin tool automatically generates specific POs for probabilistic convergence:
- PRV (Probabilistic Variant) PO: Requires proving that the variant is decreased by the probabilistic event with a positive probability. The goal is of the form
VARIANT < bound. This may require instantiating possible values for variables to demonstrate the probability is positive [93]. - BND (Bound) PO: Ensures the variant remains below the defined upper bound.
- FINACT (Finite Anticipates) PO: Ensures the set of possible new values for the variant is finite.
These POs can often be discharged automatically by the Rodin provers, but some may require interactive proof.
Case Study: Formal Verification of a Probabilistic Neural Network for Load Balancing
Recent research demonstrates the application of Event-B in verifying modern algorithms, such as an Effective Probabilistic Neural Network (EPNN) for load balancing in cloud environments [7]. This case study illustrates the integration of formal methods with machine learning.
Application Protocol and Workflow
The following diagram outlines the end-to-end workflow for developing and verifying the EPNN-based load balancer, combining machine learning training with formal modelling in Event-B.
Step 1: System Design and Modelling.
- Algorithm Design: Propose the EPNN model for selecting the optimal cluster for load distribution, alongside a Round Robin Assigning Algorithm (RRAA) for task allocation [7].
- Abstract Model Creation: Develop an initial Event-B model capturing the core system state (e.g., server nodes, request queues, load levels) and key invariants (e.g., "no single resource becomes overloaded").
Step 2: Model Refinement with EPNN Logic.
- Refine the abstract model to incorporate the EPNN's decision-making process. This involves modelling how the neural network selects the best cluster based on current system state, effectively padding the specification with algorithmic details [7] [92].
Step 3: Formal Verification.
- The Rodin tool generates proof obligations for consistency and refinement correctness. The verification process in this study involved a combination of automated and manual proof generation to ensure the algorithm's logical correctness and adherence to specified invariants [7].
Table: Quantitative Results from EPNN Load Balancer Verification
| Verification Metric | Reported Outcome | Significance for Correctness |
|---|---|---|
| Model Invariants | Verified | Ensures system properties like fault tolerance and performance are maintained [7]. |
| Refinement Steps | Consistent | Validates that each concrete model correctly implements the abstract specification [92]. |
| Proof Obligations | Automatically & Manually Discharged | Provides mathematical evidence of the algorithm's correctness under all specified conditions [7]. |
| Algorithm Accuracy | Formally Verified | Confirms the EPNN model selects the best cluster for load distribution as intended [7]. |
Discussion: Integrating Event-B into a Broader Probabilistic V&V Framework
While Event-B provides strong guarantees of logical correctness, its native qualitative probabilistic reasoning can be complemented by other V&V methodologies to form a comprehensive assurance case, particularly for perception-based autonomous systems.
A unified V&V methodology for vision-based autonomy proposes a three-step framework: abstraction, verification, and validation [17]. Event-B excels in the verification step. In this broader context:
- Abstraction: The concrete system (e.g., with neural network perception) is over-approximated by a formal model, such as an Interval Markov Decision Process (IMDP), which accounts for uncertainty with confidence intervals [17].
- Verification: The abstract model (the IMDP) is formally verified against system-level temporal properties using a probabilistic model checker. This yields a probabilistic guarantee of safety, contingent on the model's correctness [17]. Event-B could be used to verify critical components or the overall structure of this abstract model.
- Validation: The abstract model is validated against new data from a deployment environment, providing a Bayesian measure of the model's validity in the face of distribution shifts [17]. This step provides a nested probabilistic guarantee that combines frequentist (verification) and Bayesian (validation) perspectives.
This unified approach highlights how the definitive correctness proofs from tools like Event-B can be integrated with statistical validation techniques, creating rigorous yet flexible guarantees suitable for complex, learning-enabled systems operating in uncertain environments.
Conclusion
The rigorous verification and validation of probabilistic models are no longer optional but are central to modern, efficient drug development. By embracing a 'fit-for-purpose' philosophy, leveraging a diverse toolkit of methodologies, and proactively addressing implementation challenges, organizations can significantly de-risk development pipelines. The future of probabilistic modeling is deeply intertwined with emerging AI technologies, promising more dynamic, predictive, and adaptive frameworks. This evolution will ultimately enhance decision-making, improve the probability of technical and regulatory success, and accelerate the delivery of safe and effective therapies to patients in need.
References
- 1. Advancing drug development with “Fit-for-Purpose” modeling ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12436579/]
- 2. Essentials of Model-informed Drug Development in Oncology [https://www.certara.com/blog/essentials-of-model-informed-drug-development-midd-top-down-vs-bottom-up-approaches/]
- 3. Quantitative Data Quality Assurance, Analysis and Presentation [https://pmc.ncbi.nlm.nih.gov/articles/PMC12056448/]
- 4. How modeling and simulation are reshaping regulatory ... [https://www.drugdiscoverynews.com/how-modeling-and-simulation-are-reshaping-regulatory-science-16617]
- 5. Verification and validation [https://en.wikipedia.org/wiki/Verification_and_validation]
- 6. In silico trials: Verification, validation and uncertainty ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7883933/]
- 7. Formal modelling and verification of effective probabilistic ... [https://link.springer.com/article/10.1007/s10791-025-09748-2]
- 8. Verification and Validation in Software Testing [https://www.browserstack.com/guide/verification-and-validation-in-testing]
- 9. Fit-for-purpose biomarker method validation for application ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2990602/]
- 10. VeriSIM Life's Alignment with Recent FDA Draft Guidelines [https://www.verisimlife.com/publications-blog/verisim-lifes-alignment-with-recent-fda-draft-guidelines]
- 11. Verification, analytical validation, and clinical validation (V3) [https://www.nature.com/articles/s41746-020-0260-4]
- 12. Design Verification & Validation for Medical Devices [Guide] [https://www.greenlight.guru/blog/design-verification-and-design-validation]
- 13. A probabilistic approach for building disease phenotypes ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12153169/]
- 14. Requirements Verification and Validation Planning test [https://trustedinstitute.com/concept/pmi-pba/requirements-planning-management/requirements-verification-validation-planning/]
- 15. Estimating Transition Probabilities from Published Evidence [https://pmc.ncbi.nlm.nih.gov/articles/PMC7426391/]
- 16. Transition probabilities for clinical trials: investigating ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8425457/]
- 17. Towards Unified Probabilistic Verification and Validation of ... [https://arxiv.org/html/2508.14181v1]
- 18. Development of clinical probabilistic practice in Britain ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7754822/]
- 19. Carl Liebermeister and the emergence of modern medical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8902834/]
- 20. A brief history of medical statistics and its impact on ... [https://statsepi.substack.com/p/a-brief-history-of-medical-statistics]
- 21. Life Science Analytics Market Booms USD 11.27 in 2025 ... [https://www.biospace.com/press-releases/life-science-analytics-market-booms-usd-11-27-in-2025-with-north-america-at-51-share-and-ai-driving-future-growth]
- 22. 2025 Clinical Data Trend Report [https://www.veeva.com/2025-clinical-data-trend-report/]
- 23. Model-based experimental manipulation of probabilistic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9868576/]
- 24. A probabilistic construction of model validation [https://www.sciencedirect.com/science/article/abs/pii/S0045782507005014]
- 25. Probabilistic Model Checking: Applications and Trends [https://arxiv.org/html/2509.12968v1]
- 26. Physiologically Based Pharmacokinetic ( PBPK ) Modeling : A Critical... [https://www.linkedin.com/pulse/physiologically-based-pharmacokinetic-pbpk-modeling-critical-thomas-qj3if]
- 27. Model-based Meta Analysis (MBMA) Consulting [https://www.certara.com/services/model-based-meta-analysis/]
- 28. Physiologically Based Pharmacokinetic : Modeling ... Methodology [https://pmc.ncbi.nlm.nih.gov/articles/PMC3118302/]
- 29. Advancing internal exposure and physiologically-based ... [https://www.nature.com/articles/s41370-018-0046-9]
- 30. Physiologically-based PharmacoKinetic ( PBPK ) Modeling ... Tools [https://www.certara.com/publication/physiologically-based-pharmacokinetic-pbpk-modelling-tools-how-to-fit-with-our-needs/]
- 31. What Is Population Pharmacokinetic (popPK) Analysis? [https://www.allucent.com/resources/blog/what-population-pharmacokinetic-poppk-analysis]
- 32. Introduction to Pharmacokinetic Modeling Methods - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC3636497/]
- 33. Population PK/PD modelling [https://revive.gardp.org/resource/population-pk-pd-modelling/?cf=encyclopaedia]
- 34. Applications of Model-Based Meta-Analysis in Drug ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9314311/]
- 35. for Model timed automata | checking ... probabilistic Formal Methods [https://link.springer.com/article/10.1007/s10703-012-0177-x]
- 36. Temporal Logic Model Checking [https://www.coursera.org/learn/temporal-logic-model-checking]
- 37. Temporal Logic Model Checking [https://medium.com/towards-nesy/temporal-logic-model-checking-cf05fc917688]
- 38. Concept and application of the probability of pharmacological ... [https://translational-medicine.biomedcentral.com/articles/10.1186/s12967-022-03849-y]
- 39. Track: Verification, Validation, and Testing (VVT) [https://2025.quatic.org/thematic-tracks/verification-validation-and-testing]
- 40. VMCAI 2025 - 26th International Conference on Verification ... [https://conf.researchr.org/home/VMCAI-2025]
- 41. On the Concepts, Methods, and Use of "Probability ... [https://pubmed.ncbi.nlm.nih.gov/39856804/]
- 42. Leading AI-Driven Drug Discovery Platforms [https://www.sciencedirect.com/science/article/abs/pii/S0031699725075118]
- 43. Applications of machine learning in drug discovery and ... [https://www.nature.com/articles/s41573-019-0024-5]
- 44. Machine learning applications in drug development [https://www.sciencedirect.com/science/article/pii/S2001037019303988]
- 45. MIT scientists debut a generative AI model that could ... [https://news.mit.edu/2025/mit-scientists-debut-generative-ai-model-that-could-create-molecules-addressing-hard-to-treat-diseases-1125]
- 46. A Comparative Analysis of NONMEM and AI-based Models ... [https://pubmed.ncbi.nlm.nih.gov/41254220/]
- 47. VeriProP 2025 | The Workshop on Verification of Probabilistic ... [https://veriprop.github.io/2025/]
- 48. Applications of PBPK Modeling to Estimate Drug Metabolism ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12473325/]
- 49. Advancing drug development with “Fit-for-Purpose” ... [https://link.springer.com/article/10.1007/s10928-025-09995-2]
- 50. Pharmacokinetic-Pharmacodynamic Models that Incorporate ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7050630/]
- 51. Implementation of pharmacokinetic and pharmacodynamic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4112793/]
- 52. Utilizing 505(b)(2) Regulatory Pathway for New Drug ... [https://www.drugpatentwatch.com/blog/utilizing-505b2-regulatory-pathway-for-new-drug-applications-an-overview-on-the-advanced-formulation-approach-and-challenges/?srsltid=AfmBOoqKhMF3BV5hnMmECMe4UmBZIhO5MQG6L38t2SELF3j6OvtLjKfL]
- 53. Optimized 505(b)(1) and 505(b)(2) Clinical Pharmacology ... [https://premier-research.com/perspectives/optimized-505b1-and-505b2-clinical-pharmacology-programs-to-accelerate-drug-development/]
- 54. Verification of a Probabilistic Model and Optimization in ... [https://www.mdpi.com/2076-3417/15/4/1873]
- 55. Clinical Data Analysis Software [https://www.jmp.com/en/software/clinical-data-analysis-software]
- 56. elluminate Platform - Clinical Trial & Data Management ... [https://www.eclinicalsol.com/platform/]
- 57. VMD0053 Model Calibration, Validation, and Uncertainty ... [https://verra.org/methodologies/vmd0053-model-calibration-validation-and-uncertainty-guidance-for-the-methodology-for-improved-agricultural-land-management-v2-1/]
- 58. Probabilistic Audits for Verifiable Training and Outcome... [https://openreview.net/forum?id=phXKOiXC4x]
- 59. Barriers and facilitators of health professionals in adopting ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10798074/]
- 60. Barriers and facilitators to digital health adoption: a thematic ... [https://bmcdigitalhealth.biomedcentral.com/articles/10.1186/s44247-025-00227-8]
- 61. Overcoming 3 Common Barriers to Organizational Change [https://online.hbs.edu/blog/post/organizational-barriers-to-change]
- 62. to Barriers Change (+How to Overcome Them) Organizational [https://whatfix.com/blog/barriers-to-organizational-change/]
- 63. Barriers and Enablers to the Adoption of Health ... [https://www.igminresearch.com/articles/html/igmin314]
- 64. Implementing Alternative Methods [https://www.fda.gov/science-research/advancing-alternative-methods-fda/implementing-alternative-methods]
- 65. Regulatory science strategy | European Medicines Agency ... [https://www.ema.europa.eu/en/about-us/how-we-work/regulatory-science-strategy]
- 66. Recent FDA Actions Pose Big Changes and Questions for ... [https://www.hklaw.com/en/insights/publications/2025/07/recent-fda-actions-pose-big-changes-and-questions-for-health]
- 67. FDA in Flux – October 2025 Newsletter [https://www.mintz.com/insights-center/viewpoints/2791/2025-10-16-fda-flux-october-2025-newsletter]
- 68. Physiologically Based Pharmacokinetic Modeling and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12541349/]
- 69. Guideline on assessment and reporting of mechanistic models ... [https://www.ema.europa.eu/en/guideline-assessment-reporting-mechanistic-models-used-context-model-informed-drug-development]
- 70. The 6 Data Quality Dimensions with Examples [https://www.collibra.com/blog/the-6-dimensions-of-data-quality]
- 71. The Challenge of Limited Data - Adith - The Data Guy [https://adithsreeram.medium.com/the-challenge-of-limited-data-8b152b8caa38]
- 72. Application of Bayesian approaches in drug development [https://pmc.ncbi.nlm.nih.gov/articles/PMC9931171/]
- 73. What are distributional shifts and why do they matter in ... [https://ethon.ai/what-are-distributional-shifts-and-why-do-they-matter-in-industrial-applications/]
- 74. Integrating risk minimization planning throughout the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4348129/]
- 75. Understanding and Mitigating Distribution Shifts For ... [https://arxiv.org/abs/2503.08674]
- 76. Principles of Distribution Shift (PODS) [https://icml.cc/virtual/2022/workshop/13465]
- 77. Risk assessment strategies for life sciences manufacturing [https://www.crbgroup.com/insights/risk-assessment-strategies-for-life-sciences-manufacturing]
- 78. Probabilistic Verification Techniques [https://www.emergentmind.com/topics/probabilistic-verification-techniques]
- 79. Validation Protocols for Documentation Teams [https://www.docsie.io/blog/glossary/validation-protocols/]
- 80. Survey and perspective on verification, validation, and ... [https://www.nature.com/articles/s41746-025-01447-y]
- 81. Applying Probabilistic Decision Models to Clinical Trial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5961806/]
- 82. A probabilistic modeling framework for genomic networks ... [https://www.sciencedirect.com/science/article/pii/S2667237525000207]
- 83. On the Concepts, Methods, and Use of “Probability ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11924160/]
- 84. Cross-Sectional Data Analysis [https://corporatefinanceinstitute.com/resources/data-science/cross-sectional-data-analysis/]
- 85. Cross-Sectional Data - an overview [https://www.sciencedirect.com/topics/mathematics/cross-sectional-data]
- 86. Implementation of a dynamic model updating pipeline ... [https://www.sciencedirect.com/science/article/pii/S0895435624002877]
- 87. Calendar Period Estimation of Probabilities of Transition in ... [https://www.medrxiv.org/content/10.1101/2025.05.15.25327693v1]
- 88. Calendar Period Estimation of Probabilities of Transition in ... [https://www.medrxiv.org/content/10.1101/2025.05.15.25327693v1.full-text]
- 89. Reimagining patient-reported outcomes in the age of ... [https://www.nature.com/articles/s41746-025-02006-1]
- 90. Standardizing Clinical Trials Workflow Representation in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2976698/]
- 91. Event-B.org [https://www.event-b.org/]
- 92. B-Method [https://en.wikipedia.org/wiki/B-Method]
- 93. Event-B Qualitative Probability User Guide [https://wiki.event-b.org/index.php/Event-B_Qualitative_Probability_User_Guide]